Identification of Four Immune Subtypes Characterized by Distinct Composition and Functions of Tumor Microenvironment in Intrahepatic Cholangiocarcinoma
- PMID: 31875970
- PMCID: PMC7589418
- DOI: 10.1002/hep.31092
Identification of Four Immune Subtypes Characterized by Distinct Composition and Functions of Tumor Microenvironment in Intrahepatic Cholangiocarcinoma
Abstract
Background and aims: Intrahepatic cholangiocarcinoma (ICC) is a severe malignant tumor in which the standard therapies are mostly ineffective. The biological significance of the desmoplastic tumor microenvironment (TME) of ICC has been stressed but was insufficiently taken into account in the search for classifications of ICC adapted to clinical trial design. We investigated the heterogeneous tumor stroma composition and built a TME-based classification of ICC tumors that detects potentially targetable ICC subtypes.
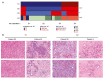
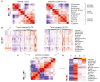
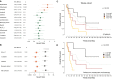
Approach and results: We established the bulk gene expression profiles of 78 ICCs. Epithelial and stromal compartments of 23 ICCs were laser microdissected. We quantified 14 gene expression signatures of the TME and those of 3 functional indicators (liver activity, inflammation, immune resistance). The cell population abundances were quantified using the microenvironment cell population-counter package and compared with immunohistochemistry. We performed an unsupervised TME-based classification of 198 ICCs (training set) and 368 ICCs (validation set). We determined immune response and signaling features of the different immune subtypes by functional annotations. We showed that a set of 198 ICCs could be classified into 4 TME-based subtypes related to distinct immune escape mechanisms and patient outcomes. The validity of these immune subtypes was confirmed over an independent set of 368 ICCs and by immunohistochemical analysis of 64 ICC tissue samples. About 45% of ICCs displayed an immune desert phenotype. The other subtypes differed in nature (lymphoid, myeloid, mesenchymal) and abundance of tumor-infiltrating cells. The inflamed subtype (11%) presented a massive T lymphocyte infiltration, an activation of inflammatory and immune checkpoint pathways, and was associated with the longest patient survival.
Conclusion: We showed the existence of an inflamed ICC subtype, which is potentially treatable with checkpoint blockade immunotherapy.
© 2020 The Authors. Hepatology published by Wiley Periodicals, Inc., on behalf of American Association for the Study of Liver Diseases.
Figures




References
-
- Poultsides GA, Zhu AX, Choti MA, Pawlik TM. Intrahepatic cholangiocarcinoma. Surg Clin North Am 2010;90:817‐837. - PubMed
-
- Valle J, Wasan H, Palmer DH, Cunningham D, Anthoney A, Maraveyas A, et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N Engl J Med 2010;362:1273‐1281. - PubMed
-
- Sirica AE. The role of cancer‐associated myofibroblasts in intrahepatic cholangiocarcinoma. Nat Rev Gastroenterol Hepatol 2011;9:44‐54. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

