Airway epithelial cell differentiation relies on deficient Hedgehog signalling in COPD
- PMID: 31877414
- PMCID: PMC6931110
- DOI: 10.1016/j.ebiom.2019.11.033
Airway epithelial cell differentiation relies on deficient Hedgehog signalling in COPD
Abstract
Background: Hedgehog (HH) pathway is constantly under scrutiny in the context of organ development. Lung morphogenesis requires HH signalling which participates thereafter to the pulmonary homeostasis by regulating epithelial cell quiescence and repair. Since epithelial remodelling is a hallmark of Chronic Obstructive Pulmonary Disease (COPD), we investigated whether the main molecular actors of HH pathway participate to airway epithelial cell differentiation and we analysed their alterations in COPD patients.
Methods: Sonic HH (Shh) secretion was assessed by ELISA in airway epithelial cell (AEC) air-liquid interface culture supernatants. HH pathway activation was evaluated by RT-qPCR, western blot and immunostaining. Inhibition of HH signalling was achieved upon Shh chelation during epithelial cell differentiation. HH pathway core components localization was investigated in lung tissues from non-COPD and COPD patients.
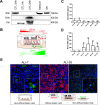
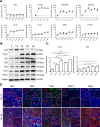
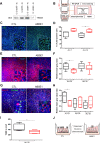
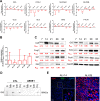
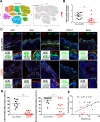
Findings: We demonstrate that progenitors of AEC produced Shh responsible for the activation of HH signalling during the process of differentiation. Preventing the ligand-induced HH activation led to the establishment of a remodelled epithelium with increased number of basal cells and reduced ciliogenesis. Gli2 activating transcription factor was demonstrated as a key-element in the regulation of AEC differentiation. More importantly, Gli2 and Smo were lost in AEC from COPD patients.
Interpretation: Our data suggest that HH pathway is crucial for airway epithelial cell differentiation and highlight its role in COPD-associated epithelial remodelling.
Keywords: Airway epithelial cells; Chronic obstructive pulmonary disease; Cilia; Differentiation; Hedgehog.
Copyright © 2019 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest Dr. Deslee reports personal fees from Nuvaira, personal fees from BTG/PneumRx, personal fees from Chiesi, personal fees from Boehringer, personal fees from Astra Zeneca, outside the submitted work.
Figures
References
-
- Singh D., Agusti A., Anzueto A., Barnes P.J., Bourbeau J., Celli B.R. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease: the gold science committee report 2019. Eur Respirat J. 2019 - PubMed
-
- Hangaard S., Helle T., Nielsen C., Hejlesen O.K. Causes of misdiagnosis of chronic obstructive pulmonary disease: a systematic scoping review. Respir Med. 2017;129:63–84. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

