Genome-Wide Analyses and Prediction of Resistance to MLN in Large Tropical Maize Germplasm
- PMID: 31877962
- PMCID: PMC7016728
- DOI: 10.3390/genes11010016
Genome-Wide Analyses and Prediction of Resistance to MLN in Large Tropical Maize Germplasm
Abstract
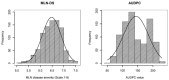
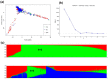
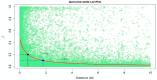
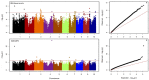
Maize lethal necrosis (MLN), caused by co-infection of maize chlorotic mottle virus and sugarcane mosaic virus, can lead up to 100% yield loss. Identification and validation of genomic regions can facilitate marker assisted breeding for resistance to MLN. Our objectives were to identify marker-trait associations using genome wide association study and assess the potential of genomic prediction for MLN resistance in a large panel of diverse maize lines. A set of 1400 diverse maize tropical inbred lines were evaluated for their response to MLN under artificial inoculation by measuring disease severity or incidence and area under disease progress curve (AUDPC). All lines were genotyped with genotyping by sequencing (GBS) SNPs. The phenotypic variation was significant for all traits and the heritability estimates were moderate to high. GWAS revealed 32 significantly associated SNPs for MLN resistance (at p < 1.0 × 10-6). For disease severity, these significantly associated SNPs individually explained 3-5% of the total phenotypic variance, whereas for AUDPC they explained 3-12% of the total proportion of phenotypic variance. Most of significant SNPs were consistent with the previous studies and assists to validate and fine map the big quantitative trait locus (QTL) regions into few markers' specific regions. A set of putative candidate genes associated with the significant markers were identified and their functions revealed to be directly or indirectly involved in plant defense responses. Genomic prediction revealed reasonable prediction accuracies. The prediction accuracies significantly increased with increasing marker densities and training population size. These results support that MLN is a complex trait controlled by few major and many minor effect genes.
Keywords: GP; GWAS; maize lethal necrosis; markers; resistance; validation.
Conflict of interest statement
The authors declare no potential conflict of interest.
Figures

References
-
- Mahuku G., Lockhart B., Wanjala B., Jones M., Kimunye J., Stewart L., Cassone B., Subramanian S., Nyasani J., Kusia E., et al. Maize lethal necrosis (MLN), an emerging threat to maize-based food security in Sub-Saharan Africa. Phytopathology. 2015;105:956–965. doi: 10.1094/PHYTO-12-14-0367-FI. - DOI - PubMed
-
- Mudde B., Olubayo F., Miano D., Asea G., Kilalo D., Kiggundu A., Kwemoi D., Adriko J. Distribution, incidence and severity of maize lethal necrosis disease in major maize growing agro-ecological zones of Uganda. J. Agric. Sci. 2018;10:72–85. doi: 10.5539/jas.v10n6p72. - DOI
-
- Adams I., Harju V., Hodges T., Hany U., Skelton A., Rai S., Deka M.K., Smith J., Fox A., Uzayisenga B., et al. First report of maize lethal necrosis disease in Rwanda. New Dis. Rep. 2014;29:22. doi: 10.5197/j.2044-0588.2014.029.022. - DOI
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

