The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor
- PMID: 31881017
- PMCID: PMC6934268
- DOI: 10.1371/journal.pgen.1008501
The MITF-SOX10 regulated long non-coding RNA DIRC3 is a melanoma tumour suppressor
Abstract
The MITF and SOX10 transcription factors regulate the expression of genes important for melanoma proliferation, invasion and metastasis. Despite growing evidence of the contribution of long noncoding RNAs (lncRNAs) in cancer, including melanoma, their functions within MITF-SOX10 transcriptional programmes remain poorly investigated. Here we identify 245 candidate melanoma associated lncRNAs whose loci are co-occupied by MITF-SOX10 and that are enriched at active enhancer-like regions. Our work suggests that one of these, Disrupted In Renal Carcinoma 3 (DIRC3), may be a clinically important MITF-SOX10 regulated tumour suppressor. DIRC3 depletion in human melanoma cells leads to increased anchorage-independent growth, a hallmark of malignant transformation, whilst melanoma patients classified by low DIRC3 expression have decreased survival. DIRC3 is a nuclear lncRNA that activates expression of its neighbouring IGFBP5 tumour suppressor through modulating chromatin structure and suppressing SOX10 binding to putative regulatory elements within the DIRC3 locus. In turn, DIRC3 dependent regulation of IGFBP5 impacts the expression of genes involved in cancer associated processes and is needed for DIRC3 control of anchorage-independent growth. Our work indicates that lncRNA components of MITF-SOX10 networks are an important new class of melanoma regulators and candidate therapeutic targets that can act not only as downstream mediators of MITF-SOX10 function but as feedback regulators of MITF-SOX10 activity.
Conflict of interest statement
The authors have declared that no competing interests exist.
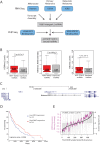
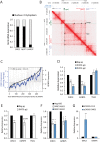
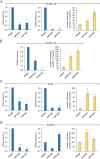
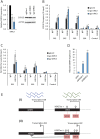
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

