Pathogen Genomics in Public Health
- PMID: 31881145
- PMCID: PMC7008580
- DOI: 10.1056/NEJMsr1813907
Pathogen Genomics in Public Health
Abstract
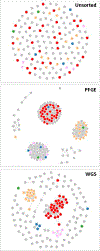
Rapid advances in DNA sequencing technology ("next-generation sequencing") have inspired optimism about the potential of human genomics for "precision medicine." Meanwhile, pathogen genomics is already delivering "precision public health" through more effective investigations of outbreaks of foodborne illnesses, better-targeted tuberculosis control, and more timely and granular influenza surveillance to inform the selection of vaccine strains. In this article, we describe how public health agencies have been adopting pathogen genomics to improve their effectiveness in almost all domains of infectious disease. This momentum is likely to continue, given the ongoing development in sequencing and sequencing-related technologies.
Copyright © 2019 Massachusetts Medical Society.
Figures
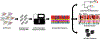
References
-
- DNA Sequencing Costs: Data. National Human Genomics Research Institute, 2018. (Accessed 08/19/2018, 2018, at https://www.genome.gov/27541954/dna-sequencing-costs-data/.)
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
