Rearrangement analysis of multiple bacterial genomes
- PMID: 31881830
- PMCID: PMC6933940
- DOI: 10.1186/s12859-019-3293-4
Rearrangement analysis of multiple bacterial genomes
Abstract
Background: Genomes are subjected to rearrangements that change the orientation and ordering of genes during evolution. The most common rearrangements that occur in uni-chromosomal genomes are inversions (or reversals) to adapt to the changing environment. Since genome rearrangements are rarer than point mutations, gene order with sequence data can facilitate more robust phylogenetic reconstruction. Helicobacter pylori is a good model because of its unique evolution in niche environment.
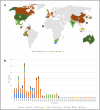
Results: We have developed a method to identify genome rearrangements by comparing almost-conserved genes among closely related strains. Orthologous gene clusters, rather than the gene sequences, are used to align the gene order so that comparison of large number of genomes becomes easier. Comparison of 72 Helicobacter pylori strains revealed shared as well as strain-specific reversals, some of which were found in different geographical locations.
Conclusion: Degree of genome rearrangements increases with time. Therefore, gene orders can be used to study the evolutionary relationship among species and strains. Multiple genome comparison helps to identify the strain-specific as well as shared reversals. Identification of the time course of rearrangements can provide insights into evolutionary events.
Keywords: Gene order; Genome rearrangements; Helicobacter pylori; Reversals.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Lara-Ramírez Edgar Eduardo, Segura-Cabrera Aldo, Guo Xianwu, Yu Gongxin, García-Pérez Carlos Armando, Rodríguez-Pérez Mario A. New Implications on Genomic Adaptation Derived from the Helicobacter pylori Genome Comparison. PLoS ONE. 2011;6(2):e17300. doi: 10.1371/journal.pone.0017300. - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

