Improvement of the memory function of a mutual repression network in a stochastic environment by negative autoregulation
- PMID: 31881978
- PMCID: PMC6935196
- DOI: 10.1186/s12859-019-3315-2
Improvement of the memory function of a mutual repression network in a stochastic environment by negative autoregulation
Abstract
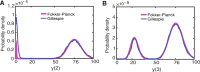
Background: Cellular memory is a ubiquitous function of biological systems. By generating a sustained response to a transient inductive stimulus, often due to bistability, memory is central to the robust control of many important biological processes. However, our understanding of the origins of cellular memory remains incomplete. Stochastic fluctuations that are inherent to most biological systems have been shown to hamper memory function. Yet, how stochasticity changes the behavior of genetic circuits is generally not clear from a deterministic analysis of the network alone. Here, we apply deterministic rate equations, stochastic simulations, and theoretical analyses of Fokker-Planck equations to investigate how intrinsic noise affects the memory function in a mutual repression network.
Results: We find that the addition of negative autoregulation improves the persistence of memory in a small gene regulatory network by reducing stochastic fluctuations. Our theoretical analyses reveal that this improved memory function stems from an increased stability of the steady states of the system. Moreover, we show how the tuning of critical network parameters can further enhance memory.
Conclusions: Our work illuminates the power of stochastic and theoretical approaches to understanding biological circuits, and the importance of considering stochasticity when designing synthetic circuits with memory function.
Keywords: Bistability; Fokker-Planck; Hill coefficient; Memory; Mutual repression; Negative autoregulation; Probability density; Stochasticity.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
MeSH terms
LinkOut - more resources
Full Text Sources

