A rapid and label-free platform for virus capture and identification from clinical samples
- PMID: 31882450
- PMCID: PMC6969489
- DOI: 10.1073/pnas.1910113117
A rapid and label-free platform for virus capture and identification from clinical samples
Abstract
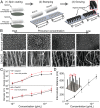
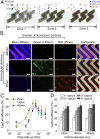
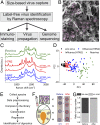
Emerging and reemerging viruses are responsible for a number of recent epidemic outbreaks. A crucial step in predicting and controlling outbreaks is the timely and accurate characterization of emerging virus strains. We present a portable microfluidic platform containing carbon nanotube arrays with differential filtration porosity for the rapid enrichment and optical identification of viruses. Different emerging strains (or unknown viruses) can be enriched and identified in real time through a multivirus capture component in conjunction with surface-enhanced Raman spectroscopy. More importantly, after viral capture and detection on a chip, viruses remain viable and get purified in a microdevice that permits subsequent in-depth characterizations by various conventional methods. We validated this platform using different subtypes of avian influenza A viruses and human samples with respiratory infections. This technology successfully enriched rhinovirus, influenza virus, and parainfluenza viruses, and maintained the stoichiometric viral proportions when the samples contained more than one type of virus, thus emulating coinfection. Viral capture and detection took only a few minutes with a 70-fold enrichment enhancement; detection could be achieved with as little as 102 EID50/mL (50% egg infective dose per microliter), with a virus specificity of 90%. After enrichment using the device, we demonstrated by sequencing that the abundance of viral-specific reads significantly increased from 4.1 to 31.8% for parainfluenza and from 0.08 to 0.44% for influenza virus. This enrichment method coupled to Raman virus identification constitutes an innovative system that could be used to quickly track and monitor viral outbreaks in real time.
Keywords: carbon nanotube; infectious disease; microfabrication; sequencing; virus isolation.
Copyright © 2020 the Author(s). Published by PNAS.
Conflict of interest statement
Competing interest statement: The authors declare no competing interest.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

