The spectraplakin Dystonin antagonizes YAP activity and suppresses tumourigenesis
- PMID: 31882643
- PMCID: PMC6934804
- DOI: 10.1038/s41598-019-56296-z
The spectraplakin Dystonin antagonizes YAP activity and suppresses tumourigenesis
Abstract
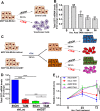
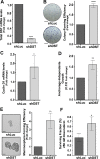
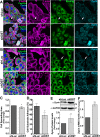
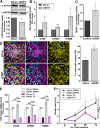
Aberrant expression of the Spectraplakin Dystonin (DST) has been observed in various cancers, including those of the breast. However, little is known about its role in carcinogenesis. In this report, we demonstrate that Dystonin is a candidate tumour suppressor in breast cancer and provide an underlying molecular mechanism. We show that in MCF10A cells, Dystonin is necessary to restrain cell growth, anchorage-independent growth, self-renewal properties and resistance to doxorubicin. Strikingly, while Dystonin maintains focal adhesion integrity, promotes cell spreading and cell-substratum adhesion, it prevents Zyxin accumulation, stabilizes LATS and restricts YAP activation. Moreover, treating DST-depleted MCF10A cells with the YAP inhibitor Verteporfin prevents their growth. In vivo, the Drosophila Dystonin Short stop also restricts tissue growth by limiting Yorkie activity. As the two Dystonin isoforms BPAG1eA and BPAG1e are necessary to inhibit the acquisition of transformed features and are both downregulated in breast tumour samples and in MCF10A cells with conditional induction of the Src proto-oncogene, they could function as the predominant Dystonin tumour suppressor variants in breast epithelial cells. Thus, their loss could deem as promising prognostic biomarkers for breast cancer.
Conflict of interest statement
All authors of this manuscript declare that they have no conflict of interest and no competing financial interests in relation to the work described. The material of this work is original research, has not been previously published and has not been submitted for publication elsewhere while under consideration.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

