ezTrack: An open-source video analysis pipeline for the investigation of animal behavior
- PMID: 31882950
- PMCID: PMC6934800
- DOI: 10.1038/s41598-019-56408-9
ezTrack: An open-source video analysis pipeline for the investigation of animal behavior
Abstract
Tracking animal behavior by video is one of the most common tasks in the life sciences. Although commercial software exists for executing this task, they often present enormous cost to the researcher and can entail purchasing hardware that is expensive and lacks adaptability. Additionally, the underlying code is often proprietary. Alternatively, available open-source options frequently require model training and can be challenging for those inexperienced with programming. Here we present an open-source and platform independent set of behavior analysis pipelines using interactive Python that researchers with no prior programming experience can use. Two modules are described. One module can be used for the positional analysis of an individual animal, amenable to a wide range of behavioral tasks. A second module is described for the analysis of freezing behavior. For both modules, a range of interactive plots and visualizations are available to confirm that chosen parameters produce the anticipated results. Moreover, batch processing tools for the fast analysis of multiple videos is provided, and frame-by-frame output makes alignment with biological recording data simple. Lastly, options for cropping video frames to mitigate the influence of fiberoptic/electrophysiology cables, analyzing specified portions of time, and defining regions of interest, are readily implemented.
Conflict of interest statement
The authors declare no competing interests.
Figures

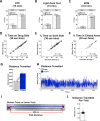

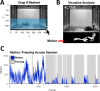

References
-
- Perez F, Granger BE. A System for Interactive Scientific Computing. Computing in Science & Engineering. 2007;9:21–29. doi: 10.1109/MCSE.2007.53. - DOI
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

