A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity
- PMID: 31883794
- PMCID: PMC7556703
- DOI: 10.1016/j.cell.2019.11.036
A Patient-Derived Glioblastoma Organoid Model and Biobank Recapitulates Inter- and Intra-tumoral Heterogeneity
Abstract
Glioblastomas exhibit vast inter- and intra-tumoral heterogeneity, complicating the development of effective therapeutic strategies. Current in vitro models are limited in preserving the cellular and mutational diversity of parental tumors and require a prolonged generation time. Here, we report methods for generating and biobanking patient-derived glioblastoma organoids (GBOs) that recapitulate the histological features, cellular diversity, gene expression, and mutational profiles of their corresponding parental tumors. GBOs can be generated quickly with high reliability and exhibit rapid, aggressive infiltration when transplanted into adult rodent brains. We further demonstrate the utility of GBOs to test personalized therapies by correlating GBO mutational profiles with responses to specific drugs and by modeling chimeric antigen receptor T cell immunotherapy. Our studies show that GBOs maintain many key features of glioblastomas and can be rapidly deployed to investigate patient-specific treatment strategies. Additionally, our live biobank establishes a rich resource for basic and translational glioblastoma research.
Keywords: CAR-T cells; Organoid; biobank; cancer modeling; drug testing; glioblastoma; personalized therapies; translational; tumor heterogeneity; xenograft.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
DECLARATION OF INTERESTS
J.F.D. has ownership interest (including equity, patents, etc.) in Liquid Biotech USA, Inc., and PolyAurum, LLC. The University of Pennsylvania has submitted a patent application based on the EGFRvIII digital PCR assay utilized. Other authors declare no competing interests.
Figures
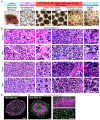
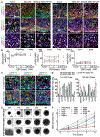


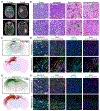
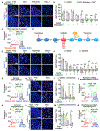

Comment in
-
Tackling the Many Facets of Glioblastoma Heterogeneity.Cell Stem Cell. 2020 Mar 5;26(3):303-304. doi: 10.1016/j.stem.2020.02.005. Cell Stem Cell. 2020. PMID: 32142659
-
Organoids: A Platform Ready for Glioblastoma Precision Medicine?Trends Cancer. 2020 Apr;6(4):265-267. doi: 10.1016/j.trecan.2020.01.016. Epub 2020 Feb 6. Trends Cancer. 2020. PMID: 32209439
References
-
- Arif SH, Pandith AA, Tabasum R, Ramzan AU, Singh S, Siddiqi MA, and Bhat AR (2018). Significant Effect of Anti-tyrosine Kinase Inhibitor (Gefitinib) on Overall Survival of the Glioblastoma Multiforme Patients in the Backdrop of Mutational Status of Epidermal Growth Factor Receptor and PTEN Genes. Asian J. Neurosurg 13, 46–52. - PMC - PubMed
-
- Backos DS, Franklin CC, and Reigan P (2012). The role of glutathione in brain tumor drug resistance. Biochem. Pharmacol 83, 1005–1012. - PubMed
-
- Bagley SJ, Schwab RD, Nelson E,Viaene AN, Binder ZA, Lustig RA, O’Rourke DM, Brem S, Desai AS, and Nasrallah MP (2019). Histopathologic quantification of viable tumor versus treatment effect in surgically resected recurrent glioblastoma. J. Neurooncol 141, 421–429. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

