Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression
- PMID: 31883952
- PMCID: PMC7033561
- DOI: 10.1016/j.molcel.2019.11.021
Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression
Abstract
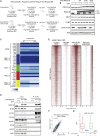
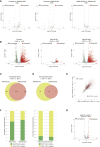
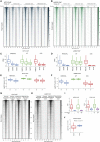
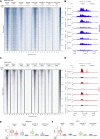
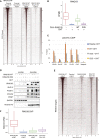
Polycomb group proteins (PcGs) maintain transcriptional repression to preserve cellular identity in two distinct repressive complexes, PRC1 and PRC2, that modify histones by depositing H2AK119ub1 and H3K27me3, respectively. PRC1 and PRC2 exist in different variants and show a complex regulatory cross-talk. However, the contribution that H2AK119ub1 plays in mediating PcG repressive functions remains largely controversial. Using a fully catalytic inactive RING1B mutant, we demonstrated that H2AK119ub1 deposition is essential to maintain PcG-target gene repression in embryonic stem cells (ESCs). Loss of H2AK119ub1 induced a rapid displacement of PRC2 activity and a loss of H3K27me3 deposition. This preferentially affected PRC2.2 variant with respect to PRC2.1, destabilizing canonical PRC1 activity. Finally, we found that variant PRC1 forms can sense H2AK119ub1 deposition, which contributes to their stabilization specifically at sites where this modification is highly enriched. Overall, our data place H2AK119ub1 deposition as a central hub that mounts PcG repressive machineries to preserve cell transcriptional identity.
Keywords: Chromatin modifications; H2AK119ub1; H3K27me3; JARID2; MTF2; PRC1; PRC2; Polycomb; RING1B; transcriptional repression.
Copyright © 2019 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declarations of Interests The authors declare no competing interests.
Figures


Comment in
-
If You Like It Then You Shoulda Put Two "RINGs" on It: Delineating the Roles of vPRC1 and cPRC1.Mol Cell. 2020 Feb 20;77(4):685-687. doi: 10.1016/j.molcel.2020.02.002. Mol Cell. 2020. PMID: 32084351
References
-
- Ben-Saadon R., Zaaroor D., Ziv T., Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol. Cell. 2006;24:701–711. - PubMed
-
- Beringer M., Pisano P., Di Carlo V., Blanco E., Chammas P., Vizán P., Gutiérrez A., Aranda S., Payer B., Wierer M., Di Croce L. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol. Cell. 2016;64:645–658. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

