The unity and diversity of the ciliary central apparatus
- PMID: 31884923
- PMCID: PMC7017334
- DOI: 10.1098/rstb.2019.0164
The unity and diversity of the ciliary central apparatus
Abstract
Nearly all motile cilia and flagella (terms here used interchangeably) have a '9+2' axoneme containing nine outer doublet microtubules and two central microtubules. The central pair of microtubules plus associated projections, termed the central apparatus (CA), is involved in the control of flagellar motility and is essential for the normal movement of '9+2' cilia. Research using the green alga Chlamydomonas reinhardtii, an important model system for studying cilia, has provided most of our knowledge of the protein composition of the CA, and recent work using this organism has expanded the number of known and candidate CA proteins nearly threefold. Here we take advantage of this enhanced proteome to examine the genomes of a wide range of eukaryotic organisms, representing all of the major phylogenetic groups, to identify predicted orthologues of the C. reinhardtii CA proteins and explore how widely the proteins are conserved and whether there are patterns to this conservation. We also discuss in detail two contrasting groups of CA proteins-the ASH-domain proteins, which are broadly conserved, and the PAS proteins, which are restricted primarily to the volvocalean algae. This article is part of the Theo Murphy meeting issue 'Unity and diversity of cilia in locomotion and transport'.
Keywords: ASH-domain proteins; Chlamydomonas; PAS-domain proteins; axoneme evolution; central microtubules; flagella.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
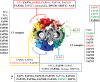
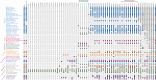
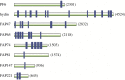

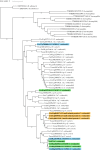
References
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
