The CTLH Complex in Cancer Cell Plasticity
- PMID: 31885576
- PMCID: PMC6907057
- DOI: 10.1155/2019/4216750
The CTLH Complex in Cancer Cell Plasticity
Abstract
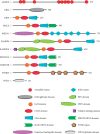
Cancer cell plasticity is the ability of cancer cells to intermittently morph into different fittest phenotypic states. Due to the intrinsic capacity to change their composition and interactions, protein macromolecular complexes are the ideal instruments for transient transformation. This review focuses on a poorly studied mammalian macromolecular complex called the CTLH (carboxy-terminal to LisH) complex. Currently, this macrostructure includes 11 known members (ARMC8, GID4, GID8, MAEA, MKLN1, RMND5A, RMND5B, RANBP9, RANBP10, WDR26, and YPEL5) and it has been shown to have E3-ligase enzymatic activity. CTLH proteins have been linked to all fundamental biological processes including proliferation, survival, programmed cell death, cell adhesion, and migration. At molecular level, the complex seems to interact and intertwine with key signaling pathways such as the PI3-kinase, WNT, TGFβ, and NFκB, which are key to cancer cell plasticity. As a whole, the CTLH complex is overexpressed in the most prevalent types of cancer and may hold the key to unlock many of the biological secrets that allow cancer cells to thrive in harsh conditions and resist antineoplastic therapy.
Copyright © 2019 Nickelas Huffman et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest.
Figures
References
-
- Sheng S., Margarida Bernardo M., Dzinic S. H., Chen K., Heath E. I., Sakr W. A. Tackling tumor heterogeneity and phenotypic plasticity in cancer precision medicine: our experience and a literature review. Cancer and Metastasis Reviews. 2018;37(4):655–663. doi: 10.1007/s10555-018-9767-4. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

