Evolution of larval segment position across 12 Drosophila species
- PMID: 31886902
- PMCID: PMC7496318
- DOI: 10.1111/evo.13911
Evolution of larval segment position across 12 Drosophila species
Abstract
Many developmental traits that are critical to the survival of the organism are also robust. These robust traits are resistant to phenotypic change in the face of variation. This presents a challenge to evolution. In this article, we asked whether and how a well-established robust trait, Drosophila segment patterning, changed over the evolutionary history of the genus. We compared segment position scaled to body length at the first-instar larval stage among 12 Drosophila species. We found that relative segment position has changed many times across the phylogeny. Changes were frequent, but primarily small in magnitude. Phylogenetic analysis demonstrated that rates of change in segment position are variable along the Drosophila phylogenetic tree, and that these changes can occur in short evolutionary timescales. Correlation between position shifts of segments decreased as the distance between two segments increased, suggesting local control of segment position. The posterior-most abdominal segment showed the highest magnitude of change on average, had the highest rate of evolution between species, and appeared to be evolving more independently as compared to the rest of the segments. This segment was exceptionally elongated in the cactophilic species in our dataset, raising questions as to whether this change may be adaptive.
Keywords: Drosophila; evolution; larval stage; robustness; segment patterning.
© 2019 The Authors. Evolution published by Wiley Periodicals, Inc. on behalf of The Society for the Study of Evolution.
Conflict of interest statement
The authors declare no competing interests.
Figures

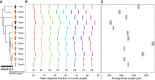
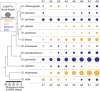


Comment in
-
Digest: Evolution of a robust phenotype in Drosophila.Evolution. 2020 Jul;74(7):1573-1574. doi: 10.1111/evo.13997. Epub 2020 May 27. Evolution. 2020. PMID: 32383154
References
-
- Arthur, W. , and Chipman A. D.. 2005. The centipede Strigamia maritima: what it can tell us about the development and evolution of segmentation. Bioessays 27:653–660. - PubMed
-
- Busturia, A. , and Lawrence P. A.. 1994. Regulation of cell number in Drosophila . Nature 370:561–563. - PubMed
-
- Clark, A. G. , Eisen M. B., Smith D. R., Bergman C. M., Oliver B., Markow T. A., Kaufman T. C., Kellis M., Gelbart W., Iyer V. N., et al. 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450:203–218. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
