Novel RNA viruses associated with Plasmodium vivax in human malaria and Leucocytozoon parasites in avian disease
- PMID: 31887217
- PMCID: PMC6953888
- DOI: 10.1371/journal.ppat.1008216
Novel RNA viruses associated with Plasmodium vivax in human malaria and Leucocytozoon parasites in avian disease
Abstract
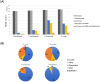
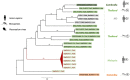
Eukaryotes of the genus Plasmodium cause malaria, a parasitic disease responsible for substantial morbidity and mortality in humans. Yet, the nature and abundance of any viruses carried by these divergent eukaryotic parasites is unknown. We investigated the Plasmodium virome by performing a meta-transcriptomic analysis of blood samples taken from patients suffering from malaria and infected with P. vivax, P. falciparum or P. knowlesi. This resulted in the identification of a narnavirus-like sequence, encoding an RNA polymerase and restricted to P. vivax samples, as well as an associated viral segment of unknown function. These data, confirmed by PCR, are indicative of a novel RNA virus that we term Matryoshka RNA virus 1 (MaRNAV-1) to reflect its analogy to a "Russian doll": a virus, infecting a parasite, infecting an animal. Additional screening revealed that MaRNAV-1 was abundant in geographically diverse P. vivax derived from humans and mosquitoes, strongly supporting its association with this parasite, and not in any of the other Plasmodium samples analyzed here nor Anopheles mosquitoes in the absence of Plasmodium. Notably, related bi-segmented narnavirus-like sequences (MaRNAV-2) were retrieved from Australian birds infected with a Leucocytozoon-a genus of eukaryotic parasites that group with Plasmodium in the Apicomplexa subclass hematozoa. Together, these data support the establishment of two new phylogenetically divergent and genomically distinct viral species associated with protists, including the first virus likely infecting Plasmodium parasites. As well as broadening our understanding of the diversity and evolutionary history of the eukaryotic virosphere, the restriction to P. vivax may be of importance in understanding P. vivax-specific biology in humans and mosquitoes, and how viral co-infection might alter host responses at each stage of the P. vivax life-cycle.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures






References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

