BacPipe: A Rapid, User-Friendly Whole-Genome Sequencing Pipeline for Clinical Diagnostic Bacteriology
- PMID: 31887656
- PMCID: PMC6941874
- DOI: 10.1016/j.isci.2019.100769
BacPipe: A Rapid, User-Friendly Whole-Genome Sequencing Pipeline for Clinical Diagnostic Bacteriology
Abstract
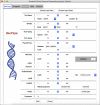
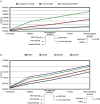
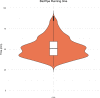
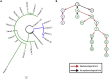
Despite rapid advances in whole genome sequencing (WGS) technologies, their integration into routine microbiological diagnostics has been hampered by the lack of standardized downstream bioinformatics analysis. We developed a comprehensive and computationally low-resource bioinformatics pipeline (BacPipe) enabling direct analyses of bacterial whole-genome sequences (raw reads or contigs) obtained from second- or third-generation sequencing technologies. A graphical user interface was developed to visualize real-time progression of the analysis. The scalability and speed of BacPipe in handling large datasets was demonstrated using 4,139 Illumina paired-end sequence files of publicly available bacterial genomes (2.9-5.4 Mb) from the European Nucleotide Archive. BacPipe is integrated in EBI-SELECTA, a project-specific portal (H2020-COMPARE), and is available as an independent docker image that can be used across Windows- and Unix-based systems. BacPipe offers a fully automated "one-stop" bacterial WGS analysis pipeline to overcome the major hurdle of WGS data analysis in hospitals and public-health and for infection control monitoring.
Keywords: Biological Sciences Research Methodologies; Microbiology; Sequence Analysis.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests None declared.
Figures





References
-
- Akgün M., Bayrak A.O., Ozer B., Sağıroğlu M.Ş. Privacy preserving processing of genomic data: a survey. J. Biomed. Inform. 2015;56:103–111. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

