Quorum Quenching Properties and Probiotic Potentials of Intestinal Associated Bacteria in Asian Sea Bass Lates calcarifer
- PMID: 31888034
- PMCID: PMC7024293
- DOI: 10.3390/md18010023
Quorum Quenching Properties and Probiotic Potentials of Intestinal Associated Bacteria in Asian Sea Bass Lates calcarifer
Abstract
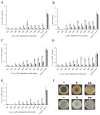
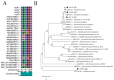
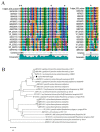
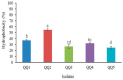
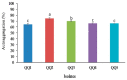
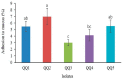
Quorum quenching (QQ), the enzymatic degradation of N-acyl homoserine lactones (AHLs), has been suggested as a promising strategy to control bacterial diseases. In this study, 10 AHL-degrading bacteria isolated from the intestine of barramundi were identified by 16S rDNA sequencing. They were able to degrade both short and long-chain AHLs associated with several pathogenic Vibrio species (spp.) in fish, including N-[(RS)-3-Hydroxybutyryl]-l-homoserine lactone (3-oh-C4-HSL), N-Hexanoyl-l-homoserine lactone (C6-HSL), N-(β-Ketocaproyl)-l-homoserine lactone (3-oxo-C6-HSL), N-(3-Oxodecanoyl)-l-homoserine lactone (3-oxo-C10-HSL), N-(3-Oxotetradecanoyl)-l-homoserine lactone (3-oxo-C14-HSL). Five QQ isolates (QQIs) belonging to the Bacillus and Shewanella genera, showed high capacity to degrade both synthetic AHLs as well as natural AHLs produced by Vibrio harveyi and Vibrio alginolyticus using the well-diffusion method and thin-layer chromatography (TLC). The genes responsible for QQ activity, including aiiA, ytnP, and aaC were also detected. Analysis of the amino acid sequences from the predicted lactonases revealed the presence of the conserved motif HxHxDH. The selected isolates were further characterized in terms of their probiotic potentials in vitro. Based on our scoring system, Bacillus thuringiensis QQ1 and Bacillus cereus QQ2 exhibited suitable probiotic characteristics, including the production of spore and exoenzymes, resistance to bile salts and pH, high potential to adhere on mucus, appropriate growth abilities, safety to barramundi, and sensitivity to antibiotics. These isolates, therefore, constitute new QQ probiotics that could be used to control vibriosis in Lates calcalifer.
Keywords: Lates calcalifer; probiotic; quorum quenching; vibriosis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Dawood M.A., Koshio S., Ishikawa M., Yokoyama S., El Basuini M.F., Hossain M.S., Nhu T.H., Dossou S., Moss A.S. Effects of dietary supplementation of Lactobacillus rhamnosus or/and Lactococcus lactis on the growth, gut microbiota and immune responses of red sea bream, Pagrus major. Fish Shellfish. Immunol. 2016;49:275–285. doi: 10.1016/j.fsi.2015.12.047. - DOI - PubMed
-
- Novriadi R. Vibriosis in aquaculture. Omni-Akuatika. 2016:12. doi: 10.20884/1.oa.2016.12.1.24. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

