Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing
- PMID: 31888061
- PMCID: PMC7016817
- DOI: 10.3390/genes11010030
Identification of Exogenous Nitric Oxide-Responsive miRNAs from Alfalfa (Medicago sativa L.) under Drought Stress by High-Throughput Sequencing
Abstract
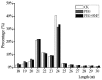
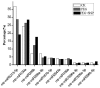
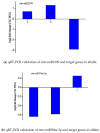
Alfalfa (Medicago sativa L.) is a high quality leguminous forage. Drought stress is one of the main factors that restrict the development of the alfalfa industry. High-throughput sequencing was used to analyze the microRNA (miRNA) profiles of alfalfa plants treated with CK (normal water), PEG (polyethylene glycol-6000; drought stress), and PEG + SNP (sodium nitroprusside; nitric oxide (NO) sprayed externally under drought stress). We identified 90 known miRNAs belonging to 46 families and predicted 177 new miRNAs. Real-time quantitative fluorescent PCR (qRT-PCR) was used to validate high-throughput expression analysis data. A total of 32 (14 known miRNAs and 18 new miRNAs) and 55 (24 known miRNAs and 31 new miRNAs) differentially expressed miRNAs were identified in PEG and PEG + SNP samples. This suggested that exogenous NO can induce more new miRNAs. The differentially expressed miRNA maturation sequences in the two treatment groups were targeted by 86 and 157 potential target genes, separately. The function of target genes was annotated by gene ontology (GO) enrichment and kyoto encyclopedia of genes and genomes (KEGG) analysis. The expression profiles of nine selected miRNAs and their target genes verified that their expression patterns were opposite. This study has documented that analysis of miRNA under PEG and PEG + SNP conditions provides important insights into the improvement of drought resistance of alfalfa by exogenous NO at the molecular level. This has important scientific value and practical significance for the improvement of plant drought resistance by exogenous NO.
Keywords: alfalfa; differential expression; drought stress; exogenous nitric oxide; miRNAs.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







Similar articles
-
Mechanism of action of microRNA166 on nitric oxide in alfalfa (Medicago sativa L.) under drought stress.BMC Genomics. 2024 Mar 28;25(1):316. doi: 10.1186/s12864-024-10095-7. BMC Genomics. 2024. PMID: 38549050 Free PMC article.
-
Regulation of endogenous hormone and miRNA in leaves of alfalfa (Medicago sativa L.) seedlings under drought stress by endogenous nitric oxide.BMC Genomics. 2024 Mar 1;25(1):229. doi: 10.1186/s12864-024-10024-8. BMC Genomics. 2024. PMID: 38429670 Free PMC article.
-
Analysis of physiological and miRNA responses to Pi deficiency in alfalfa (Medicago sativa L.).Plant Mol Biol. 2018 Mar;96(4-5):473-492. doi: 10.1007/s11103-018-0711-3. Epub 2018 Mar 12. Plant Mol Biol. 2018. PMID: 29532290
-
Unveiling the biosynthesis, mechanisms, and impacts of miRNAs in drought stress resilience in plants.Plant Physiol Biochem. 2023 Sep;202:107978. doi: 10.1016/j.plaphy.2023.107978. Epub 2023 Aug 24. Plant Physiol Biochem. 2023. PMID: 37660607 Review.
-
NO to drought-multifunctional role of nitric oxide in plant drought: Do we have all the answers?Plant Sci. 2015 Oct;239:44-55. doi: 10.1016/j.plantsci.2015.07.012. Epub 2015 Jul 23. Plant Sci. 2015. PMID: 26398790 Review.
Cited by
-
Screening of mtr-miR156a from exosomes of dairy cow blood to milk and its regulatory effect on milk protein synthesis in BMECs.BMC Genomics. 2024 Sep 19;25(1):882. doi: 10.1186/s12864-024-10761-w. BMC Genomics. 2024. PMID: 39300336 Free PMC article.
-
Mechanism of action of microRNA166 on nitric oxide in alfalfa (Medicago sativa L.) under drought stress.BMC Genomics. 2024 Mar 28;25(1):316. doi: 10.1186/s12864-024-10095-7. BMC Genomics. 2024. PMID: 38549050 Free PMC article.
-
Redox-Epigenetic Crosstalk in Plant Stress Responses: The Roles of Reactive Oxygen and Nitrogen Species in Modulating Chromatin Dynamics.Int J Mol Sci. 2025 Jul 24;26(15):7167. doi: 10.3390/ijms26157167. Int J Mol Sci. 2025. PMID: 40806301 Free PMC article. Review.
-
MicroRNAs: Potential Targets for Developing Stress-Tolerant Crops.Life (Basel). 2021 Mar 28;11(4):289. doi: 10.3390/life11040289. Life (Basel). 2021. PMID: 33800690 Free PMC article. Review.
-
MsTHI1 overexpression improves drought tolerance in transgenic alfalfa (Medicago sativa L.).Front Plant Sci. 2022 Sep 8;13:992024. doi: 10.3389/fpls.2022.992024. eCollection 2022. Front Plant Sci. 2022. PMID: 36160983 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

