GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae
- PMID: 31888478
- PMCID: PMC6937711
- DOI: 10.1186/s12870-019-2132-0
GmWRKY40, a member of the WRKY transcription factor genes identified from Glycine max L., enhanced the resistance to Phytophthora sojae
Abstract
Background: The WRKY proteins are a superfamily of transcription factors and members play essential roles in the modulation of diverse physiological processes, such as growth, development, senescence and response to biotic and abiotic stresses. However, the biological roles of the majority of the WRKY family members remains poorly understood in soybean relative to the research progress in model plants.
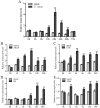
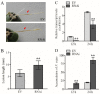
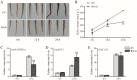
Results: In this study, we identified and characterized GmWRKY40, which is a group IIc WRKY gene. Transient expression analysis revealed that the GmWRKY40 protein is located in the nucleus of plant cells. Expression of GmWRKY40 was strongly induced in soybean following infection with Phytophthora sojae, or treatment with methyl jasmonate, ethylene, salicylic acid, and abscisic acid. Furthermore, soybean hairy roots silencing GmWRKY40 enhanced susceptibility to P. sojae infection compared with empty vector transgenic roots. Moreover, suppression of GmWRKY40 decreased the accumulation of reactive oxygen species (ROS) and modified the expression of several oxidation-related genes. Yeast two-hybrid experiment combined with RNA-seq analysis showed that GmWRKY40 interacted with 8 JAZ proteins with or without the WRKY domain or zinc-finger domain of GmWRKY40, suggesting there were different interaction patterns among these interacted proteins.
Conclusions: Collectively, these results suggests that GmWRKY40 functions as a positive regulator in soybean plants response to P. sojae through modulating hydrogen peroxide accumulation and JA signaling pathway.
Keywords: Glycine max (L.) Merr; GmWRKY40; Phytophthora sojae; RNA interference; Soybean hairy roots; Yeast two-hybrid.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Sugimoto T, Kato M, Yoshida S, Matsumoto I, Kobayashi T, Kaga A, Hajika M, Yamamoto R, Watanabe K, Aino M, et al. Pathogenic diversity of Phytophthora sojae and breeding strategies to develop Phytophthora-resistant soybeans. Breed Sci. 2012;61(5):511–522. doi: 10.1270/jsbbs.61.511. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- 2017YFD0101500/National Key R&D Program of China
- KYZ201811/Fundamental Research Funds for the Central Universities
- 2016ZX08004002/Genetically Modified Organisms Breeding Major Projects
- CARS-04-PS10/Modern Agro-industry Technology Research System of China
- PCSIRT_17R55/Program for Changjiang Scholars and Innovative Research Team in University
LinkOut - more resources
Full Text Sources

