Nitric oxide- induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana
- PMID: 31888479
- PMCID: PMC6937950
- DOI: 10.1186/s12870-019-2210-3
Nitric oxide- induced AtAO3 differentially regulates plant defense and drought tolerance in Arabidopsis thaliana
Abstract
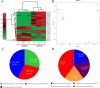
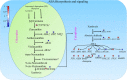
Background: Exposure of plants to different environmental insults instigates significant changes in the cellular redox tone driven in part by promoting the production of reactive nitrogen species. The key player, nitric oxide (NO) is a small gaseous diatomic molecule, well-known for its signaling role during stress. In this study, we focused on abscisic acid (ABA) metabolism-related genes that showed differential expression in response to the NO donor S-nitroso-L-cysteine (CySNO) by conducting RNA-seq-based transcriptomic analysis.
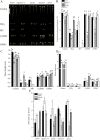
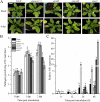
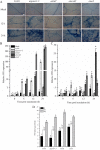
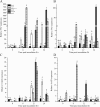
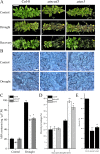
Results: CySNO-induced ABA-related genes were identified and further characterized. Gene ontology terms for biological processes showed most of the genes were associated with protein phosphorylation. Promoter analysis suggested that several cis-regulatory elements were activated under biotic and/or abiotic stress conditions. The ABA biosynthetic gene AtAO3 was selected for validation using functional genomics. The loss of function mutant atao3 was found to differentially regulate oxidative and nitrosative stress. Further investigations for determining the role of AtAO3 in plant defense suggested a negative regulation of plant basal defense and R-gene-mediated resistance. The atao3 plants showed resistance to virulent Pseudomonas syringae pv. tomato strain DC3000 (Pst DC3000) with gradual increase in PR1 gene expression. Similarly, atao3 plants showed increased hypersensitive response (HR) when challenged with Pst DC3000 (avrB). The atgsnor1-3 and atsid2 mutants showed a susceptible phenotype with reduced PR1 transcript accumulation. Drought tolerance assay indicated that atao3 and atnced3 ABA-deficient mutants showed early wilting, followed by plant death. The study of stomatal structure showed that atao3 and atnced3 were unable to close stomata even at 7 days after drought stress. Further, they showed reduced ABA content and increased electrolyte leakage than the wild-type (WT) plants. The quantitative polymerase chain reaction analysis suggested that ABA biosynthesis genes were down-regulated, whereas expression of most of the drought-related genes were up-regulated in atao3 than in WT.
Conclusions: AtAO3 negatively regulates pathogen-induced salicylic acid pathway, although it is required for drought tolerance, despite the fact that ABA production is not totally dependent on AtAO3, and that drought-related genes like DREB2 and ABI2 show response to drought irrespective of ABA content.
Keywords: ABA metabolism genes; Basal defense; Drought stress; Nitrosative stress; R-gene-mediated resistance; RNA-seq analysis; Stomatal regulation.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

