OHMI: the ontology of host-microbiome interactions
- PMID: 31888755
- PMCID: PMC6937947
- DOI: 10.1186/s13326-019-0217-1
OHMI: the ontology of host-microbiome interactions
Abstract
Background: Host-microbiome interactions (HMIs) are critical for the modulation of biological processes and are associated with several diseases. Extensive HMI studies have generated large amounts of data. We propose that the logical representation of the knowledge derived from these data and the standardized representation of experimental variables and processes can foster integration of data and reproducibility of experiments and thereby further HMI knowledge discovery.
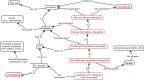
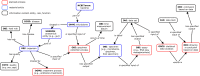
Methods: Through a multi-institutional collaboration, a community-based Ontology of Host-Microbiome Interactions (OHMI) was developed following the Open Biological/Biomedical Ontologies (OBO) Foundry principles. As an OBO library ontology, OHMI leverages established ontologies to create logically structured representations of (1) microbiomes, microbial taxonomy, host species, host anatomical entities, and HMIs under different conditions and (2) associated study protocols and types of data analysis and experimental results.
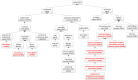
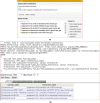
Results: Aligned with the Basic Formal Ontology, OHMI comprises over 1000 terms, including terms imported from more than 10 existing ontologies together with some 500 OHMI-specific terms. A specific OHMI design pattern was generated to represent typical host-microbiome interaction studies. As one major OHMI use case, drawing on data from over 50 peer-reviewed publications, we identified over 100 bacteria and fungi from the gut, oral cavity, skin, and airway that are associated with six rheumatic diseases including rheumatoid arthritis. Our ontological study identified new high-level microbiota taxonomical structures. Two microbiome-related competency questions were also designed and addressed. We were also able to use OHMI to represent statistically significant results identified from a large existing microbiome database data analysis.
Conclusion: OHMI represents entities and relations in the domain of HMIs. It supports shared knowledge representation, data and metadata standardization and integration, and can be used in formulation of advanced queries for purposes of data analysis.
Keywords: Host-microbiome interaction; Metadata; Microbiome; OBO Foundry; OHMI; Ontology; Ontology of host-microbiome interactions; Rheumatic disease; Rheumatoid arthritis.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

