Moran Model of Spatial Alignment in Microbial Colonies
- PMID: 31889737
- PMCID: PMC6936756
- DOI: 10.1016/j.physd.2019.02.001
Moran Model of Spatial Alignment in Microbial Colonies
Abstract
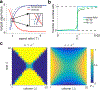
We describe a spatial Moran model that captures mechanical interactions and directional growth in spatially extended populations. The model is analytically tractable and completely solvable under a mean-field approximation and can elucidate the mechanisms that drive the formation of population-level patterns. As an example we model a population of E. coli growing in a rectangular microfluidic trap. We show that spatial patterns can arise as a result of a tug-of-war between boundary effects and growth rate modulations due to cell-cell interactions: Cells align parallel to the long side of the trap when boundary effects dominate. However, when cell-cell interactions exceed a critical value, cells align orthogonally to the trap's long side. This modeling approach and analysis can be extended to directionally-growing cells in a variety of domains to provide insight into how local and global interactions shape collective behavior.
Keywords: Moran model; cell alignment; mean field; phase transition.
Figures




References
-
- Parrish JK, Viscido SV, and Grünbaum D, “Self-organized fish schools: An examination of emergent properties,” Biol. Bull 202 (2002). - PubMed
-
- Chaté H, Ginelli F, Grégoire G, Peruani F, and Reynaud F, “Modeling collective motion: variations on the vicsek model,” Euro. Phys. J. B 64, 451–456 (2008).
-
- Peruani F, Starruß J, Jakovljevic V, Søgaard-Andersen L, Deutsch A, and Bär M, “Collective motion and nonequilibrium cluster formation in colonies of gliding bacteria,” Phys. Rev. Lett 108 (12). - PubMed
-
- Murray JD, “A pre-pattern formation mechanism for animal coat markings,” J. Theor. Biol 99, 161–199 (1981).
-
- Murray JD, “How the leopard got its stripes,” Scientific American 258, 80–87 (1988).
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
