Isothermal titration calorimetry and surface plasmon resonance analysis using the dynamic approach
- PMID: 31890903
- PMCID: PMC6926116
- DOI: 10.1016/j.bbrep.2019.100712
Isothermal titration calorimetry and surface plasmon resonance analysis using the dynamic approach
Erratum in
-
Corrigendum "Isothermal titration calorimetry and surface plasmon resonance analysis using the dynamic approach" [Biochem. Biophys. Rep. 21 (2020) 100712].Biochem Biophys Rep. 2020 Jan 7;22:100723. doi: 10.1016/j.bbrep.2019.100723. eCollection 2020 Jul. Biochem Biophys Rep. 2020. PMID: 32490212 Free PMC article.
-
Corrigendum to "Isothermal Titration Calorimetry and Surface Plasmon Resonance Analysis Using the Dynamic Approach".Biochem Biophys Rep. 2020 Oct 3;24:100800. doi: 10.1016/j.bbrep.2020.100800. eCollection 2020 Dec. Biochem Biophys Rep. 2020. PMID: 33381662 Free PMC article.
Abstract
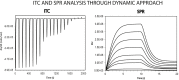
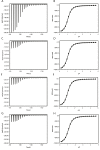
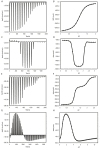
Biophysical techniques such as isothermal titration calorimetry (ITC) and surface plasmon resonance (SPR) are routinely used to ascertain the global binding mechanisms of protein-protein or protein-ligand interaction. Recently, Dumas etal, have explicitly modelled the instrument response of the ligand dilution and analysed the ITC thermogram to obtain kinetic rate constants. Adopting a similar approach, we have integrated the dynamic instrument response with the binding mechanism to simulate the ITC profiles of equivalent and independent binding sites, equivalent and sequential binding sites and aggregating systems. The results were benchmarked against the standard commercial software Origin-ITC. Further, the experimental ITC chromatograms of 2'-CMP + RNASE and BH3I-1 + hBCLXL interactions were analysed and shown to be comparable with that of the conventional analysis. Dynamic approach was applied to simulate the SPR profiles of a two-state model, and could reproduce the experimental profile accurately.
Keywords: Aggregation model; BH3I-1; Dynamic approach; Equivalent binding; ITC; Instrument response; RNASE; SPR; Sequential binding; hBCLXL.
© 2019 The Authors.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
-
- Martinez J., Murciano-Calles J., Iglesias-Bexiga E., Luque I., Ruiz-Sanz J. Appl. Calorim. A Wide Context-Differ. Scanning Calorimetry, Isothermal Titration Calorim. Microcalorim. vols 73–104. 2013. Isothermal titration calorimetry: thermodynamic analysis of the binding thermograms of molecular recognition events by using equilibrium models. (There is no corresponding record for this reference.[Google Scholar])
-
- Jelesarov I., Bosshard H.R. Isothermal titration calorimetry and differential scanning calorimetry as complementary tools to investigate the energetics of biomolecular recognition. J. Mol. Recognit. 1999;12:3–18. - PubMed
-
- Pierce M.M., Raman C., Nall B.T. Isothermal titration calorimetry of protein–protein interactions. Methods. 1999;19:213–221. - PubMed
-
- Wiseman T., Williston S., Brandts J.F., Lin L.-N. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal. Biochem. 1989;179:131–137. - PubMed
-
- Freyer M.W., Lewis E.A. Isothermal titration calorimetry: experimental design, data analysis, and probing macromolecule/ligand binding and kinetic interactions. Methods Cell Biol. 2008;84:79–113. - PubMed
LinkOut - more resources
Full Text Sources

