Comparative analysis of eight DNA extraction methods for molecular research in mealybugs
- PMID: 31891602
- PMCID: PMC6938366
- DOI: 10.1371/journal.pone.0226818
Comparative analysis of eight DNA extraction methods for molecular research in mealybugs
Abstract
For molecular research, the quality and integrity of DNA obtained will affect the reliability of subsequent results. Extracting quality DNA from scale insects, including mealybugs, can be difficult due to their small body size and waxy coating. In this study, we evaluate eight commonly used DNA extraction methods to determine their efficacy in PCR analysis across life stages and preservation times. We find that fresh samples, immediately upon collection or after 2 wks, resulted in the most effective DNA extraction. Methods using the DNeasy Blood & Tissue kit, NaCl, SDS-RNase A, and SDS isolated DNA of sufficient quality DNA. The SDS method gave high DNA yield, while the NaCl and SDS-RNase A methods gave lower yield. NaCl, SDS-RNase A, SDS, chloroform-isopentyl alcohol, and the salting-out methods all resulted in sufficient DNA for PCR, and performed equal to or better than that of the DNeasy Blood & Tissue kit. When time and cost per extraction were considered, the SDS method was most efficient, especially for later life stages of mealybug, regardless of preservation duration. DNA extracted from a single fresh sample of a female adult mealybug was adequate for more than 10,000 PCR reactions. For earlier stages, including the egg and 1st instar nymph samples, DNA was most effectively extracted by the Rapid method. Our results provide guidelines for the choice of effective DNA extraction method for mealybug or other small insects across different life stages and preservation status.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
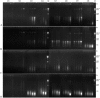

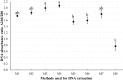

References
-
- Hardy NB, Gullan PJ, Hodgson CJ. A subfamily-level classification of mealybugs (Hemiptera: Pseudococcidae) based on integrated molecular and morphological data. Syst Entomol. 2008;33: 51–71.
-
- ScaleNet. Pseudococcidae; 2016. [cited 26 April 2019]. http://scalenet.info/fams/Pseudococcidae.
-
- Zeddies J, Schaab RP, Neuenschwander P, Herren HR. Economics of biological control of cassava mealybug in Africa. Agr Econ. 2001;24: 209–219.
-
- Miller DR, Miller GL, Watson GW. Invasive species of mealybugs (Hemiptera: Pseudococcidae) and their threat to US agriculture. Proc Entomol Soc Wash. 2002;104: 825–836.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

