A phenome-wide association study (PheWAS) in the Population Architecture using Genomics and Epidemiology (PAGE) study reveals potential pleiotropy in African Americans
- PMID: 31891604
- PMCID: PMC6938343
- DOI: 10.1371/journal.pone.0226771
A phenome-wide association study (PheWAS) in the Population Architecture using Genomics and Epidemiology (PAGE) study reveals potential pleiotropy in African Americans
Abstract
We performed a hypothesis-generating phenome-wide association study (PheWAS) to identify and characterize cross-phenotype associations, where one SNP is associated with two or more phenotypes, between thousands of genetic variants assayed on the Metabochip and hundreds of phenotypes in 5,897 African Americans as part of the Population Architecture using Genomics and Epidemiology (PAGE) I study. The PAGE I study was a National Human Genome Research Institute-funded collaboration of four study sites accessing diverse epidemiologic studies genotyped on the Metabochip, a custom genotyping chip that has dense coverage of regions in the genome previously associated with cardio-metabolic traits and outcomes in mostly European-descent populations. Here we focus on identifying novel phenome-genome relationships, where SNPs are associated with more than one phenotype. To do this, we performed a PheWAS, testing each SNP on the Metabochip for an association with up to 273 phenotypes in the participating PAGE I study sites. We identified 133 putative pleiotropic variants, defined as SNPs associated at an empirically derived p-value threshold of p<0.01 in two or more PAGE study sites for two or more phenotype classes. We further annotated these PheWAS-identified variants using publicly available functional data and local genetic ancestry. Amongst our novel findings is SPARC rs4958487, associated with increased glucose levels and hypertension. SPARC has been implicated in the pathogenesis of diabetes and is also known to have a potential role in fibrosis, a common consequence of multiple conditions including hypertension. The SPARC example and others highlight the potential that PheWAS approaches have in improving our understanding of complex disease architecture by identifying novel relationships between genetic variants and an array of common human phenotypes.
Conflict of interest statement
SAP is employed by and receives salary from Genentech Inc, and JMJ is employed by and receives salary from Illumina, Inc. This does not alter our adherence with PLOS ONE policies on sharing data and materials. DCC is an academic editor of PLOS ONE. DCC is not involved in the review of this manuscript per journal policy.
Figures
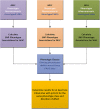
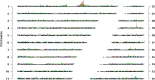




References
Publication types
MeSH terms
Grants and funding
- N01 HC055016/HL/NHLBI NIH HHS/United States
- P30 ES010126/ES/NIEHS NIH HHS/United States
- N01 HC055019/HL/NHLBI NIH HHS/United States
- UL1 TR002548/TR/NCATS NIH HHS/United States
- U01 HG004790/HG/NHGRI NIH HHS/United States
- N01 WH022110/WH/WHI NIH HHS/United States
- N01 HC055015/HL/NHLBI NIH HHS/United States
- N01 HC055020/HL/NHLBI NIH HHS/United States
- U01 HG004803/HG/NHGRI NIH HHS/United States
- N01 HC055018/HL/NHLBI NIH HHS/United States
- U01 CA098758/CA/NCI NIH HHS/United States
- N01 HC055022/HL/NHLBI NIH HHS/United States
- U01 HG004798/HG/NHGRI NIH HHS/United States
- U01 CA136792/CA/NCI NIH HHS/United States
- U01 HG004801/HG/NHGRI NIH HHS/United States
- U01 HG004802/HG/NHGRI NIH HHS/United States
- N01 HC055021/HL/NHLBI NIH HHS/United States
- P01 CA033619/CA/NCI NIH HHS/United States
- UL1 TR000439/TR/NCATS NIH HHS/United States
- P30 CA071789/CA/NCI NIH HHS/United States
- R37 CA054281/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

