Pixelated colorimetric nucleic acid assay
- PMID: 31892020
- PMCID: PMC7111824
- DOI: 10.1016/j.talanta.2019.120581
Pixelated colorimetric nucleic acid assay
Abstract
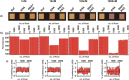
Conjugated polyelectrolytes (CPEs) have been widely used as reporters in colorimetric assays targeting nucleic acids. CPEs provide naked eye detection possibility by their superior optical properties however, as concentration of target analytes decrease, trace amounts of nucleic acid typically yield colorimetric responses that are not readily perceivable by naked eye. Herein, we report a pixelated analysis approach for correlating colorimetric responses of CPE with nucleic acid concentrations down to 1 nM, in plasma samples, utilizing a smart phone with an algorithm that can perform analytical testing and data processing. The detection strategy employed relies on conformational transitions between single stranded nucleic acid-cationic CPE duplexes and double stranded nucleic acid-CPE triplexes that yield distinct colorimetric responses for enabling naked eye detection of nucleic acids. Cationic poly[N,N,N-triethyl-3-((4-methylthiophen-3-yl)oxy)propan-1-aminium bromide] is utilized as the CPE reporter deposited on a polyvinylidene fluoride (PVDF) membrane for nucleic acid assay. A smart phone application is developed to capture and digitize the colorimetric response of the individual pixels of the digital images of CPE on the PVDF membrane, followed by an analysis using the algorithm. The proposed pixelated approach enables precise quantification of nucleic acid assay concentrations, thereby eliminating the margin of error involved in conventional methodologies adopted for interpretation of colorimetric responses, for instance, RGB analysis. The obtained results illustrate that a ubiquitous smart phone could be utilized for point of care colorimetric nucleic acids assays in complex matrices without requiring sophisticated software or instrumentation.
Keywords: Conjugated polyelectrolyte; Diagnostic tool; Nucleic acid assay; Paper-based sensor; Pixelated analysis; Point-of-care.
Copyright © 2019 Elsevier B.V. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures





References
-
- Baeumner A.J., Schlesinger N.A., Slutzki N.S., Romano J., Lee E.M., Montagna R.A. Biosensor for dengue virus Detection: sensitive, rapid, and serotype specific. Anal. Chem. 2002;74(6):1442–1448. - PubMed
-
- Choi J.R., Tang R., Wang S., Wan Abas W.A., Pingguan-Murphy B., Xu F. Paper-based sample-to-answer molecular diagnostic platform for point-of-care diagnostics. Biosens. Bioelectron. 2015;74:427–439. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

