Folding Rate Optimization Promotes Frustrated Interactions in Entangled Protein Structures
- PMID: 31892272
- PMCID: PMC6981561
- DOI: 10.3390/ijms21010213
Folding Rate Optimization Promotes Frustrated Interactions in Entangled Protein Structures
Abstract
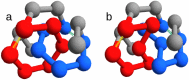
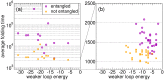
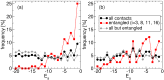
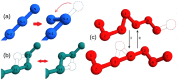
Many native structures of proteins accomodate complex topological motifs such as knots, lassos, and other geometrical entanglements. How proteins can fold quickly even in the presence of such topological obstacles is a debated question in structural biology. Recently, the hypothesis that energetic frustration might be a mechanism to avoid topological frustration has been put forward based on the empirical observation that loops involved in entanglements are stabilized by weak interactions between amino-acids at their extrema. To verify this idea, we use a toy lattice model for the folding of proteins into two almost identical structures, one entangled and one not. As expected, the folding time is longer when random sequences folds into the entangled structure. This holds also under an evolutionary pressure simulated by optimizing the folding time. It turns out that optmized protein sequences in the entangled structure are in fact characterized by frustrated interactions at the closures of entangled loops. This phenomenon is much less enhanced in the control case where the entanglement is not present. Our findings, which are in agreement with experimental observations, corroborate the idea that an evolutionary pressure shapes the folding funnel to avoid topological and kinetic traps.
Keywords: entanglement; protein folding; topological frustration.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Folding kinetics of an entangled protein.PLoS Comput Biol. 2023 Nov 13;19(11):e1011107. doi: 10.1371/journal.pcbi.1011107. eCollection 2023 Nov. PLoS Comput Biol. 2023. PMID: 37956216 Free PMC article.
-
Sequence and structural patterns detected in entangled proteins reveal the importance of co-translational folding.Sci Rep. 2019 Jun 10;9(1):8426. doi: 10.1038/s41598-019-44928-3. Sci Rep. 2019. PMID: 31182755 Free PMC article.
-
On folding of entangled proteins: knots, lassos, links and θ-curves.Curr Opin Struct Biol. 2020 Feb;60:131-141. doi: 10.1016/j.sbi.2020.01.007. Epub 2020 Feb 12. Curr Opin Struct Biol. 2020. PMID: 32062143 Review.
-
Folding pathway dependence on energetic frustration and interaction heterogeneity for a three-dimensional hydrophobic protein model.Proteins. 2006 Jan 1;62(1):46-63. doi: 10.1002/prot.20711. Proteins. 2006. PMID: 16292745
-
Fuzziness and Frustration in the Energy Landscape of Protein Folding, Function, and Assembly.Acc Chem Res. 2021 Mar 2;54(5):1251-1259. doi: 10.1021/acs.accounts.0c00813. Epub 2021 Feb 8. Acc Chem Res. 2021. PMID: 33550810 Free PMC article. Review.
Cited by
-
Folding kinetics of an entangled protein.PLoS Comput Biol. 2023 Nov 13;19(11):e1011107. doi: 10.1371/journal.pcbi.1011107. eCollection 2023 Nov. PLoS Comput Biol. 2023. PMID: 37956216 Free PMC article.
-
Entangled Motifs in Membrane Protein Structures.Int J Mol Sci. 2023 May 24;24(11):9193. doi: 10.3390/ijms24119193. Int J Mol Sci. 2023. PMID: 37298146 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

