Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework
- PMID: 31892348
- PMCID: PMC6938631
- DOI: 10.1186/s13073-019-0690-2
Recommendations for application of the functional evidence PS3/BS3 criterion using the ACMG/AMP sequence variant interpretation framework
Abstract
Background: The American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) clinical variant interpretation guidelines established criteria for different types of evidence. This includes the strong evidence codes PS3 and BS3 for "well-established" functional assays demonstrating a variant has abnormal or normal gene/protein function, respectively. However, they did not provide detailed guidance on how functional evidence should be evaluated, and differences in the application of the PS3/BS3 codes are a contributor to variant interpretation discordance between laboratories. This recommendation seeks to provide a more structured approach to the assessment of functional assays for variant interpretation and guidance on the use of various levels of strength based on assay validation.
Methods: The Clinical Genome Resource (ClinGen) Sequence Variant Interpretation (SVI) Working Group used curated functional evidence from ClinGen Variant Curation Expert Panel-developed rule specifications and expert opinions to refine the PS3/BS3 criteria over multiple in-person and virtual meetings. We estimated the odds of pathogenicity for assays using various numbers of variant controls to determine the minimum controls required to reach moderate level evidence. Feedback from the ClinGen Steering Committee and outside experts were incorporated into the recommendations at multiple stages of development.
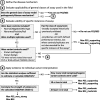
Results: The SVI Working Group developed recommendations for evaluators regarding the assessment of the clinical validity of functional data and a four-step provisional framework to determine the appropriate strength of evidence that can be applied in clinical variant interpretation. These steps are as follows: (1) define the disease mechanism, (2) evaluate the applicability of general classes of assays used in the field, (3) evaluate the validity of specific instances of assays, and (4) apply evidence to individual variant interpretation. We found that a minimum of 11 total pathogenic and benign variant controls are required to reach moderate-level evidence in the absence of rigorous statistical analysis.
Conclusions: The recommendations and approach to functional evidence evaluation described here should help clarify the clinical variant interpretation process for functional assays. Further, we hope that these recommendations will help develop productive partnerships with basic scientists who have developed functional assays that are useful for interrogating the function of a variety of genes.
Keywords: Functional assays; Guidelines; Variant interpretation.
Conflict of interest statement
LGB is an unpaid consultant of Illumina and received in-kind research support from ArQule Inc., and honoraria from Cold Spring Harbor Press. The remaining authors declare that they have no competing interests.
Figures
References
-
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. - DOI - PMC - PubMed
-
- Amendola LM, Jarvik GP, Leo MC, McLaughlin HM, Akkari Y, Amaral MD, et al. Performance of ACMG-AMP variant-interpretation guidelines among nine laboratories in the Clinical Sequencing Exploratory Research Consortium. Am J Hum Genet. 2016;98:1067–1076. doi: 10.1016/j.ajhg.2016.03.024. - DOI - PMC - PubMed
-
- Sequence Variant Interpretation Working Group [Internet]. [cited 2019 Oct 16]. Available from: https://www.clinicalgenome.org/working-groups/sequence-variant-interpret.... Accessed 16 Oct 2019.
Publication types
MeSH terms
Grants and funding
- R01 DE023414/DE/NIDCR NIH HHS/United States
- 5T32 GM007092/GM/NIGMS NIH HHS/United States
- R01 CA121245/CA/NCI NIH HHS/United States
- U41 HG006834/HG/NHGRI NIH HHS/United States
- U01 CA116167/CA/NCI NIH HHS/United States
- R01 CA225262/CA/NCI NIH HHS/United States
- U41 HG009649/HG/NHGRI NIH HHS/United States
- R01 DK044003/DK/NIDDK NIH HHS/United States
- U41HG006834/HG/NHGRI NIH HHS/United States
- 5T32 GM008719-6/GM/NIGMS NIH HHS/United States
- R01 CA222477/CA/NCI NIH HHS/United States
- ZIA HG200359 10/HG/NHGRI NIH HHS/United States
- 3U41HG009650-02S1/HG/NHGRI NIH HHS/United States
- P50 CA116201/CA/NCI NIH HHS/United States
- R01 CA116167/CA/NCI NIH HHS/United States
- U41HG009649/HG/NHGRI NIH HHS/United States
- U41 HG009650/HG/NHGRI NIH HHS/United States
- T32 GM008719/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

