In silico approach to the analysis of SNPs in the human APAF1 gene
- PMID: 31892812
- PMCID: PMC6911258
- DOI: 10.3906/biy-1905-18
In silico approach to the analysis of SNPs in the human APAF1 gene
Abstract
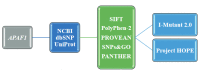
The apoptotic protease activating factor 1 (APAF1) gene encodes a cytoplasmic protein that initiates apoptosis and is a crucial factor in the mitochondria-dependent death pathway. APAF1 is implicated in many pathways such as apoptosis, neurodegenerative diseases, and cancer. The purpose of this study was to predict deleterious/damaging SNPs in the APAF1 gene viain silicoanalysis. To this end, APAF1 missense SNPs were obtained from the NCBI dbSNP database. In silico analysis of the missense SNPs was carried out by using publicly available online software tools. The stabilization and three-dimensional modeling of mutant proteins were also determined by using the I-Mutant 2.0 and Project HOPE webservers, respectively. In total, 772 missense SNPs were found in the APAF1 gene from the NCBI dbSNP database, 18 SNPs of which were demonstrated to be deleterious or damaging. Of those, 13 SNPs had a decreasing effect on protein stability, while the other 5 SNPs had an increasing effect. Based on the modeling results, some dissimilarities of mutant type amino acids from wild-type amino acids such as size, charge, and hydrophobicity were revealed. The SNPs predicted to be deleterious in this study might be used in the selection of target SNPs for genotyping in disease association studies. Therefore, we could suggest that the present study could pave the way for future experimental studies.
Keywords: APAF1; apoptosis; in silico; neurodegenerative diseases; single nucleotide polymorphism (SNP).
Copyright © 2019 The Author(s).
Conflict of interest statement
CONFLICT OF INTEREST: none declared
Figures

References
-
- Acehan D Jiang X Morgan DG Heuser JE Wang X Three-dimensional structure of the apoptosome: implications for assembly, procaspase-9 binding, and activation. Molecular Cell. 2002;9:423. - PubMed
-
- Ashkenazi A Salvesen G Regulated cell death: signaling and mechanisms. Annual Review of Cell and Developmental Biology. 2014;30:337. - PubMed
-
- Bao Q Lu W Rabinowitz JD Shi Y Calcium blocks formation of apoptosome by preventing nucleotide exchange in Apaf-1. Molecular Cell. 2007;25:181. - PubMed
LinkOut - more resources
Full Text Sources
