Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway spruce
- PMID: 31892945
- PMCID: PMC6935592
- DOI: 10.1111/eva.12823
Multi-trait genomic selection for weevil resistance, growth, and wood quality in Norway spruce
Abstract
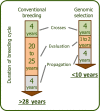
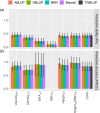
Plantation-grown trees have to cope with an increasing pressure of pest and disease in the context of climate change, and breeding approaches using genomics may offer efficient and flexible tools to face this pressure. In the present study, we targeted genetic improvement of resistance of an introduced conifer species in Canada, Norway spruce (Picea abies (L.) Karst.), to the native white pine weevil (Pissodes strobi Peck). We developed single- and multi-trait genomic selection (GS) models and selection indices considering the relationships between weevil resistance, intrinsic wood quality, and growth traits. Weevil resistance, acoustic velocity as a proxy for mechanical wood stiffness, and average wood density showed moderate-to-high heritability and low genotype-by-environment interactions. Weevil resistance was genetically positively correlated with tree height, height-to-diameter at breast height (DBH) ratio, and acoustic velocity. The accuracy of the different GS models tested (GBLUP, threshold GBLUP, Bayesian ridge regression, BayesCπ) was high and did not differ among each other. Multi-trait models performed similarly as single-trait models when all trees were phenotyped. However, when weevil attack data were not available for all trees, weevil resistance was more accurately predicted by integrating genetically correlated growth traits into multi-trait GS models. A GS index that corresponded to the breeders' priorities achieved near maximum gains for weevil resistance, acoustic velocity, and height growth, but a small decrease for DBH. The results of this study indicate that it is possible to breed for high-quality, weevil-resistant Norway spruce reforestation stock with high accuracy achieved from single-trait or multi-trait GS.
Keywords: Norway spruce; breeding; conifers; index selection; insect resistance; multi‐trait genomic selection; white pine weevil; wood quality.
© 2019 Her Majesty the Queen in Right of Canada. Environmental DNA published by John Wiley & Sons Ltd.
Figures


References
-
- Alfaro, R. I. , King, J. N. , & VanAkker, L. (2013). Delivering Sitka spruce with resistance against white pine weevil in British Columbia, Canada. The Forestry Chronicle, 89(2), 235–245. 10.5558/tfc2013-042 - DOI
-
- Alfaro, R. I. , VanAkker, L. , Jaquish, B. , & King, J. (2004). Weevil resistance of progeny derived from putatively resistant and susceptible interior spruce parents. Forest Ecology and Management, 202(1–3), 369–377. 10.1016/j.foreco.2004.08.001 - DOI
-
- Aubry, C. A. , Adams, W. T. , & Fahey, T. D. (1998). Determination of relative economic weights for multitrait selection in coastal Douglas‐fir. Canadian Journal of Forest Research, 28(8), 1164–1170. 10.1139/x98-084 - DOI
-
- Azaiez, A. , Pavy, N. , Gérardi, S. , Laroche, J. , Boyle, B. , Gagnon, F. , … Bousquet, J. (2018). A catalog of annotated high‐confidence SNPs from exome capture and sequencing reveals highly polymorphic genes in Norway spruce (Picea abies). BMC Genomics, 19, 942 10.1186/s12864-018-5247-z - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

