Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery
- PMID: 31894140
- PMCID: PMC7078311
- DOI: 10.1038/s41416-019-0672-6
Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery
Abstract
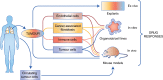
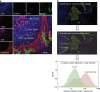
Preclinical models that can accurately predict outcomes in the clinic are much sought after in the field of cancer drug discovery and development. Existing models such as organoids and patient-derived xenografts have many advantages, but they suffer from the drawback of not contextually preserving human tumour architecture. This is a particular problem for the preclinical testing of immunotherapies, as these agents require an intact tumour human-specific microenvironment for them to be effective. In this review, we explore the potential of patient-derived explants (PDEs) for fulfilling this need. PDEs involve the ex vivo culture of fragments of freshly resected human tumours that retain the histological features of original tumours. PDE methodology for anti-cancer drug testing has been in existence for many years, but the platform has not been widely adopted in translational research facilities, despite strong evidence for its clinical predictivity. By modifying PDE endpoint analysis to include the spatial profiling of key biomarkers by using multispectral imaging, we argue that PDEs offer many advantages, including the ability to correlate drug responses with tumour pathology, tumour heterogeneity and changes in the tumour microenvironment. As such, PDEs are a powerful model of choice for cancer drug and biomarker discovery programmes.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

