Human Tumor-Lymphatic Microfluidic Model Reveals Differential Conditioning of Lymphatic Vessels by Breast Cancer Cells
- PMID: 31894641
- PMCID: PMC7004876
- DOI: 10.1002/adhm.201900925
Human Tumor-Lymphatic Microfluidic Model Reveals Differential Conditioning of Lymphatic Vessels by Breast Cancer Cells
Abstract
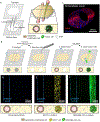
Breast tumor progression is a complex process involving intricate crosstalk between the primary tumor and its microenvironment. In the context of breast tumor-lymphatic interactions, it is unclear how breast cancer cells alter the gene expression of lymphatic endothelial cells and how these transcriptional changes potentiate lymphatic dysfunction. Thus, there is a need for in vitro lymphatic vessel models to study these interactions. In this work, a tumor-lymphatic microfluidic model is developed to study the differential conditioning of lymphatic vessels by estrogen receptor-positive (i.e., MCF7) and triple-negative (i.e., MDA-MB-231) breast cancer cells. The model consists of a lymphatic endothelial vessel cultured adjacently to either MCF7 or MDA-MB-231 cells. Quantitative transcriptional analysis reveals expression changes in genes related to vessel growth, permeability, metabolism, hypoxia, and apoptosis in lymphatic endothelial cells cocultured with breast cancer cells. Interestingly, these changes are different in the MCF7-lymphatic cocultures as compared to the 231-lymphatic cocultures. Importantly, these changes in gene expression correlate to functional responses, such as endothelial barrier dysfunction. These results collectively demonstrate the utility of this model for studying breast tumor-lymphatic crosstalk for multiple breast cancer subtypes.
Keywords: breast cancer; estrogen receptor-positive; lymphatic; microfluidic; triple-negative.
© 2019 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Conflict of interest statement
Conflict of Interest
David J. Beebe is a board member and stockowner of Tasso, Inc. and a stockowner of Bellbrook Labs, LLC. David J. Beebe is a founder, stockowner, and consultant of Salus Discovery LLC. David J. Beebe is an advisor and stockowner of Lynx Biosciences, LLC, Onexio Biosystems, LLC, and Stacks to the Future, LLC. David J. Beebe holds equity in Bellbrook Labs, LLC, Tasso Inc., Salus Discovery LLC, Stacks to the Future, LLC and Onexio Biosystems, LLC.
Figures



References
-
- Tang C, Wang P, Li X, Zhao B, Yang H, Yu H, Li C, Plos One 2017, 12, e0182953; - PMC - PubMed
- Truong PT, Vinh-Hung V, Cserni G, Woodward WA, Tai P, Vlastos G, Eur J Cancer 2008, 44, 1670; - PubMed
- Shen J, Hunt KK, Mirza NQ, Krishnamurthy S, Singletary SE, Kuerer HM, Meric-Bernstam F, Feig B, Ross MI, Ames FC, Babiera GV, Cancer 2004, 101, 1330. - PubMed
-
- Alitalo A, Detmar M, Oncogene 2012, 31, 4499. - PubMed
Publication types
MeSH terms
Grants and funding
- Morgridge Research Institute/International
- R01 CA205101/CA/NCI NIH HHS/United States
- W81XWH-13-1-0194/Department of Defense Breast Cancer Research Program/International
- P50 DE026787/DE/NIDCR NIH HHS/United States
- 067-16/Mary Kay Foundation/International
- R01 CA186134/CA/NCI NIH HHS/United States
- R01 CA185747/NH/NIH HHS/United States
- R01 CA164492/CA/NCI NIH HHS/United States
- R01 CA164492/NH/NIH HHS/United States
- R01 CA205101/NH/NIH HHS/United States
- CBET-1642287/National Science Foundation/International
- R01 CA185747/CA/NCI NIH HHS/United States
- AAB7173/University of Wisconsin Carbone Cancer Center/International
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

