Imaging FCS delineates subtle heterogeneity in plasma membranes of resting mast cells
- PMID: 31895009
- PMCID: PMC7202073
- DOI: 10.1091/mbc.E19-10-0559
Imaging FCS delineates subtle heterogeneity in plasma membranes of resting mast cells
Abstract
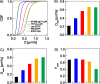
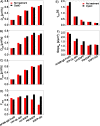
A myriad of transient, nanoscopic lipid- and protein-based interactions confer a steady-state organization of the plasma membrane in resting cells that is poised to orchestrate assembly of key signaling components upon reception of an extracellular stimulus. Although difficult to observe directly in live cells, these subtle interactions can be discerned by their impact on the diffusion of membrane constituents. Here, we quantified the diffusion properties of a panel of structurally distinct lipid, lipid-anchored, and transmembrane (TM) probes in RBL mast cells by imaging fluorescence correlation spectroscopy (ImFCS). We developed a statistical analysis of data combined from many pixels over multiple cells to characterize differences in diffusion coefficients as small as 10%, which reflect differences in underlying interactions. We found that the distinctive diffusion properties of lipid probes can be explained by their dynamic partitioning into Lo-like proteolipid nanodomains, which encompass a major fraction of the membrane and whose physical properties are influenced by actin polymerization. Effects on diffusion of functional protein modules in both lipid--anchored and TM probes reflect additional complexity in steady state membrane organization. The contrast we observe between different probes diffusing through the same membrane milieu represents the dynamic resting steady state, which serves as a baseline for monitoring plasma membrane remodeling that occurs upon stimulation.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

