Alternate RNA Structures
- PMID: 31896543
- PMCID: PMC6942119
- DOI: 10.1101/cshperspect.a032425
Alternate RNA Structures
Abstract
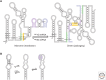
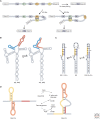
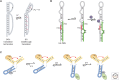
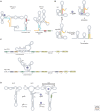
RNA molecules fold into complex three-dimensional structures that sample alternate conformations ranging from minor differences in tertiary structure dynamics to major differences in secondary structure. This allows them to form entirely different substructures with each population potentially giving rise to a distinct biological outcome. The substructures can be partitioned along an existing energy landscape given a particular static cellular cue or can be shifted in response to dynamic cues such as ligand binding. We review a few key examples of RNA molecules that sample alternate conformations and how these are capitalized on for control of critical regulatory functions.
Copyright © 2020 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures
Similar articles
-
Mapping the landscape of RNA dynamics with NMR spectroscopy.Acc Chem Res. 2011 Dec 20;44(12):1292-301. doi: 10.1021/ar200137d. Epub 2011 Sep 6. Acc Chem Res. 2011. PMID: 21894962
-
HIV-1 spliced RNAs display transcription start site bias.RNA. 2020 Jun;26(6):708-714. doi: 10.1261/rna.073650.119. Epub 2020 Mar 23. RNA. 2020. PMID: 32205324 Free PMC article.
-
The molecular interactions that stabilize RNA tertiary structure: RNA motifs, patterns, and networks.Acc Chem Res. 2011 Dec 20;44(12):1302-11. doi: 10.1021/ar200098t. Epub 2011 Sep 7. Acc Chem Res. 2011. PMID: 21899297
-
Structure and function of the human immunodeficiency virus leader RNA.Prog Nucleic Acid Res Mol Biol. 1996;54:1-34. doi: 10.1016/s0079-6603(08)60359-1. Prog Nucleic Acid Res Mol Biol. 1996. PMID: 8768071 Review. No abstract available.
-
Modulation of RNA function by oligonucleotides recognizing RNA structure.Prog Nucleic Acid Res Mol Biol. 2001;69:1-46. doi: 10.1016/s0079-6603(01)69043-3. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11550792 Review.
Cited by
-
Molecular and Immunological Properties of a Chimeric Glycosyl Hydrolase 18 Based on Immunoinformatics Approaches: A Design of a New Anti-Leishmania Vaccine.ACS Pharmacol Transl Sci. 2024 Dec 31;8(1):78-96. doi: 10.1021/acsptsci.4c00341. eCollection 2025 Jan 10. ACS Pharmacol Transl Sci. 2024. PMID: 39816796
-
Intrinsic Regulatory Role of RNA Structural Arrangement in Alternative Splicing Control.Int J Mol Sci. 2020 Jul 21;21(14):5161. doi: 10.3390/ijms21145161. Int J Mol Sci. 2020. PMID: 32708277 Free PMC article. Review.
-
TAS-seq enables subcellular single-stranded adenosine profiling by signal peptide-assisted adenosine deamination.Cell Rep Methods. 2025 Jul 21;5(7):101087. doi: 10.1016/j.crmeth.2025.101087. Epub 2025 Jun 25. Cell Rep Methods. 2025. PMID: 40570838 Free PMC article.
-
e-RNA: a collection of web-servers for the prediction and visualisation of RNA secondary structure and their functional features.Nucleic Acids Res. 2023 Jul 5;51(W1):W160-W167. doi: 10.1093/nar/gkad296. Nucleic Acids Res. 2023. PMID: 37099369 Free PMC article.
-
Length-dependent motions of SARS-CoV-2 frameshifting RNA pseudoknot and alternative conformations suggest avenues for frameshifting suppression.Res Sq [Preprint]. 2022 Jan 4:rs.3.rs-1160075. doi: 10.21203/rs.3.rs-1160075/v1. Res Sq. 2022. Update in: Nat Commun. 2022 Jul 25;13(1):4284. doi: 10.1038/s41467-022-31353-w. PMID: 35018371 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
