Molecular and clonal evolution in recurrent metastatic gliosarcoma
- PMID: 31896544
- PMCID: PMC6996521
- DOI: 10.1101/mcs.a004671
Molecular and clonal evolution in recurrent metastatic gliosarcoma
Abstract
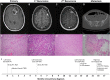
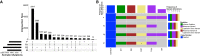
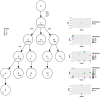
We discuss the molecular evolution of gliosarcoma, a mesenchymal type of glioblastoma (GBM), using the case of a 37-yr-old woman who developed two recurrences and an extracranial metastasis. She was initially diagnosed with isocitrate dehydrogenase (IDH) wild-type gliosarcoma in the frontal lobe and treated with surgery followed by concurrent radiotherapy with temozolomide. Five months later the tumor recurred in the left frontal lobe, outside the initially resected area, and was treated with further surgery and radiotherapy. Six months later the patient developed a second left frontal recurrence and was again treated with surgery and radiotherapy. Six weeks later, further recurrence was observed in the brain and bone, and biopsy confirmed metastases in the pelvic bones. To understand the clonal relationships between the four tumor instances and the origin of metastasis, we performed whole-genome sequencing of the intracranial tumors and the tumor located in the right iliac bone. We compared their mutational and copy-number profiles and inferred the clonal phylogeny. The tumors harbored shared alterations in GBM driver genes, including mutations in TP53, NF1, and RB1, and CDKN2A deletion. Whole-genome doubling was identified in the first recurrence and the extracranial metastasis. Comparisons of the metastatic to intracranial tumors highlighted a high similarity in molecular profile but contrasting evidence regarding the origin of the metastasis. Subclonal reconstruction suggested a parallel evolution of the recurrent tumors, and that the metastatic tumor was largely derived from the first recurrence. We conclude that metastasis in glioma can be a late event in tumorigenesis.
Keywords: IDH; glioblastoma; glioma; neoplasm of the nervous system.
© 2020 Anderson et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Akgül S, Patch A-M, D'Souza RCJ, Mukhopadhyay P, Nones K, Kempe S, Kazakoff SH, Jeffree RL, Stringer BW, Pearson JV, et al. 2019. Intratumoural heterogeneity underlies distinct therapy responses and treatment resistance in glioblastoma. Cancers (Basel) 11: E190 10.3390/cancers11020190 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
