Phylogenetics, Genomic Recombination, and NSP2 Polymorphic Patterns of Porcine Reproductive and Respiratory Syndrome Virus in China and the United States in 2014-2018
- PMID: 31896589
- PMCID: PMC7158704
- DOI: 10.1128/JVI.01813-19
Phylogenetics, Genomic Recombination, and NSP2 Polymorphic Patterns of Porcine Reproductive and Respiratory Syndrome Virus in China and the United States in 2014-2018
Abstract
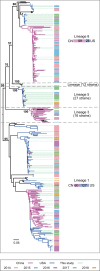
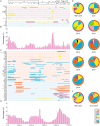
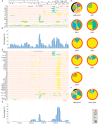
Porcine reproductive and respiratory syndrome virus (PRRSV), an important pathogen that affects the pig industry, is a highly genetically diverse RNA virus. However, the phylogenetic and genomic recombination properties of this virus have not been completely elucidated. In this study, comparative analyses of all available genomic sequences of North American (NA)-type PRRSVs (n = 355, including 138 PRRSV genomes sequenced in this study) in China and the United States during 2014-2018 revealed a high frequency of interlineage recombination hot spots in nonstructural protein 9 (NSP9) and the GP2 to GP3 regions. Lineage 1 (L1) PRRSV was found to be susceptible to recombination among PRRSVs both in China and the United States. The recombinant major parent between the 1991-2013 data and the 2014-2018 data showed a trend from complex to simple. The major recombination pattern changed from an L8 to L1 backbone during 2014-2018 for Chinese PRRSVs, whereas L1 was always the major backbone for US PRRSVs. Intralineage recombination hot spots were not as concentrated as interlineage recombination hot spots. In the two main clades with differential diversity in L1, NADC30-like PRRSVs are undergoing a decrease in population genetic diversity, NADC34-like PRRSVs have been relatively stable in population genetic diversity for years. Systematic analyses of insertion and deletion (indel) polymorphisms of NSP2 divided PRRSVs into 25 patterns, which could generate novel references for the classification of PRRSVs. The results of this study contribute to a deeper understanding of the recombination of PRRSVs and indicate the need for coordinated epidemiological investigations among countries.IMPORTANCE Porcine reproductive and respiratory syndrome (PRRS) is one of the most significant swine diseases. However, the phylogenetic and genomic recombination properties of the PRRS virus (PRRSV) have not been completely elucidated. In this study, we systematically compared differences in the lineage distribution, recombination, NSP2 polymorphisms, and evolutionary dynamics between North American (NA)-type PRRSVs in China and in the United States. Strikingly, we found high frequency of interlineage recombination hot spots in nonstructural protein 9 (NSP9) and in the GP2 to GP3 region. Also, intralineage recombination hot spots were scattered across the genome between Chinese and US strains. Furthermore, we proposed novel methods based on NSP2 indel patterns for the classification of PRRSVs. Evolutionary dynamics analysis revealed that NADC30-like PRRSVs are undergoing a decrease in population genetic diversity, suggesting that a dominant population may occur and cause an outbreak. Our findings offer important insights into the recombination of PRRSVs and suggest the need for coordinated international epidemiological investigations.
Keywords: deletion; interlineage; porcine reproductive and respiratory syndrome virus; recombination hotspots.
Copyright © 2020 American Society for Microbiology.
Figures

Similar articles
-
Analysis of Recombinant Characteristics Based on 949 PRRSV-2 Genomic Sequences Obtained from 1991 to 2021 Shows That Viral Multiplication Ability Contributes to Dominant Recombination.Microbiol Spectr. 2022 Oct 26;10(5):e0293422. doi: 10.1128/spectrum.02934-22. Epub 2022 Sep 8. Microbiol Spectr. 2022. PMID: 36073823 Free PMC article.
-
Recombination in lineage 1, 3, 5 and 8 of porcine reproductive and respiratory syndrome viruses in China.Infect Genet Evol. 2019 Mar;68:119-126. doi: 10.1016/j.meegid.2018.12.006. Epub 2018 Dec 5. Infect Genet Evol. 2019. PMID: 30529558
-
Insights into the evolutionary history and epidemiological characteristics of the emerging lineage 1 porcine reproductive and respiratory syndrome viruses in China.Transbound Emerg Dis. 2020 Nov;67(6):2630-2641. doi: 10.1111/tbed.13613. Epub 2020 Jun 5. Transbound Emerg Dis. 2020. PMID: 32386249
-
The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective.Virol J. 2018 Jan 4;15(1):2. doi: 10.1186/s12985-017-0910-6. Virol J. 2018. PMID: 29301547 Free PMC article. Review.
-
The evolution and diversity of porcine reproductive and respiratory syndrome virus in China.Vet Microbiol. 2024 Nov;298:110252. doi: 10.1016/j.vetmic.2024.110252. Epub 2024 Sep 12. Vet Microbiol. 2024. PMID: 39299012 Review.
Cited by
-
Molecular epizootiology of porcine reproductive and respiratory syndrome virus in the Xinjiang Uygur Autonomous Region of China.Front Microbiol. 2024 Jun 26;15:1419499. doi: 10.3389/fmicb.2024.1419499. eCollection 2024. Front Microbiol. 2024. PMID: 38989028 Free PMC article.
-
Reverse genetics construction and pathogenicity of a novel recombinant NADC30-like PRRSV isolated in China.Front Vet Sci. 2024 Jun 26;11:1434539. doi: 10.3389/fvets.2024.1434539. eCollection 2024. Front Vet Sci. 2024. PMID: 38993278 Free PMC article.
-
Research Progress on the NSP9 Protein of Porcine Reproductive and Respiratory Syndrome Virus.Front Vet Sci. 2022 Jul 11;9:872205. doi: 10.3389/fvets.2022.872205. eCollection 2022. Front Vet Sci. 2022. PMID: 35898550 Free PMC article. Review.
-
Genetic characterization and pathogenicity of two recombinant PRRSV-2 strains from lineages 1, 3, 5, and 8 emerged in China.BMC Vet Res. 2025 May 15;21(1):341. doi: 10.1186/s12917-025-04779-9. BMC Vet Res. 2025. PMID: 40369592 Free PMC article.
-
Lycopene Inhibits PRRSV Replication by Suppressing ROS Production.Int J Mol Sci. 2025 Aug 5;26(15):7560. doi: 10.3390/ijms26157560. Int J Mol Sci. 2025. PMID: 40806687 Free PMC article.
References
-
- Collins JE, Benfield DA, Christianson WT, Harris L, Hennings JC, Shaw DP, Goyal SM, McCullough S, Morrison RB, Joo HS. 1992. Isolation of swine infertility and respiratory syndrome virus (isolate ATCC VR-2332) in North America and experimental reproduction of the disease in gnotobiotic pigs. J Vet Diagn Invest 4:117–126. doi:10.1177/104063879200400201. - DOI - PubMed
-
- Neumann EJ, Kliebenstein JB, Johnson CD, Mabry JW, Bush EJ, Seitzinger AH, Green AL, Zimmerman JJ. 2005. Assessment of the economic impact of porcine reproductive and respiratory syndrome on swine production in the United States. J Am Vet Med Assoc 227:385–392. doi:10.2460/javma.2005.227.385. - DOI - PubMed
-
- Tian K, Yu X, Zhao T, Feng Y, Cao Z, Wang C, Hu Y, Chen X, Hu D, Tian X, Liu D, Zhang S, Deng X, Ding Y, Yang L, Zhang Y, Xiao H, Qiao M, Wang B, Hou L, Wang X, Yang X, Kang L, Sun M, Jin P, Wang S, Kitamura Y, Yan J, Gao GF. 2007. Emergence of fatal PRRSV variants: unparalleled outbreaks of atypical PRRS in China and molecular dissection of the unique hallmark. PLoS One 2:e526. doi:10.1371/journal.pone.0000526. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

