Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion
- PMID: 31896772
- PMCID: PMC6940358
- DOI: 10.1038/s41467-019-13867-y
Genome-scale reconstructions of the mammalian secretory pathway predict metabolic costs and limitations of protein secretion
Abstract
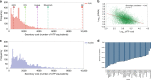
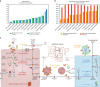
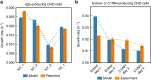
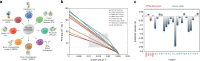
In mammalian cells, >25% of synthesized proteins are exported through the secretory pathway. The pathway complexity, however, obfuscates its impact on the secretion of different proteins. Unraveling its impact on diverse proteins is particularly important for biopharmaceutical production. Here we delineate the core secretory pathway functions and integrate them with genome-scale metabolic reconstructions of human, mouse, and Chinese hamster ovary cells. The resulting reconstructions enable the computation of energetic costs and machinery demands of each secreted protein. By integrating additional omics data, we find that highly secretory cells have adapted to reduce expression and secretion of other expensive host cell proteins. Furthermore, we predict metabolic costs and maximum productivities of biotherapeutic proteins and identify protein features that most significantly impact protein secretion. Finally, the model successfully predicts the increase in secretion of a monoclonal antibody after silencing a highly expressed selection marker. This work represents a knowledgebase of the mammalian secretory pathway that serves as a novel tool for systems biotechnology.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Uhlen M., Fagerberg L., Hallstrom B. M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., Olsson I., Edlund K., Lundberg E., Navani S., Szigyarto C. A.-K., Odeberg J., Djureinovic D., Takanen J. O., Hober S., Alm T., Edqvist P.-H., Berling H., Tegel H., Mulder J., Rockberg J., Nilsson P., Schwenk J. M., Hamsten M., von Feilitzen K., Forsberg M., Persson L., Johansson F., Zwahlen M., von Heijne G., Nielsen J., Ponten F. Tissue-based map of the human proteome. Science. 2015;347(6220):1260419–1260419. doi: 10.1126/science.1260419. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

