Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics
- PMID: 31896784
- PMCID: PMC7082340
- DOI: 10.1038/s41396-019-0571-0
Novel syntrophic bacteria in full-scale anaerobic digesters revealed by genome-centric metatranscriptomics
Abstract
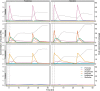
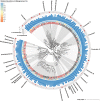
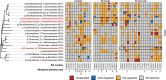
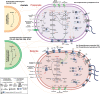
Short-chain fatty acid (SCFA) degradation is an important process in methanogenic ecosystems, and is usually catalyzed by SCFA-oxidizing bacteria in syntrophy with methanogens. Current knowledge of this functional guild is mainly based on isolates or enrichment cultures, but these may not reflect the true diversity and in situ activities of the syntrophs predominating in full-scale systems. Here we obtained 182 medium to high quality metagenome-assembled genomes (MAGs) from the microbiome of two full-scale anaerobic digesters. The transcriptomic response of individual MAG was studied after stimulation with low concentrations of acetate, propionate, or butyrate, separately. The most pronounced response to butyrate was observed for two MAGs of the recently described genus Candidatus Phosphitivorax (phylum Desulfobacterota), expressing a butyrate beta-oxidation pathway. For propionate, the largest response was observed for an MAG of a novel genus in the family Pelotomaculaceae, transcribing a methylmalonyl-CoA pathway. All three species were common in anaerobic digesters at Danish wastewater treatment plants as shown by amplicon analysis, and this is the first time their syntrophic features involved in SCFA oxidation were revealed with transcriptomic evidence. Further, they also possessed unique genomic features undescribed in well-characterized syntrophs, including the metabolic pathways for phosphite oxidation, nitrite and sulfate reduction.
Conflict of interest statement
MD, PHN, RHK, MA and TYM are employed by DNASense ApS. The other authors declare no conflict of interest.
Figures
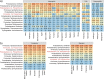
References
-
- McInerney MJ, Sieber JR, Gunsalus RP. Microbial syntrophy: ecosystem-level biochemical cooperation. Microbe Mag. 2011;6:479–85. doi: 10.1128/microbe.6.479.1. - DOI
-
- Schink B, Stams AJM. Syntrophism among Prokaryotes. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Berlin, Heidelberg: Springer Berlin Heidelberg; 2013. pp. 471–93.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

