The role of deubiquitinating enzymes in gastric cancer
- PMID: 31897112
- PMCID: PMC6924028
- DOI: 10.3892/ol.2019.11062
The role of deubiquitinating enzymes in gastric cancer
Abstract
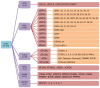
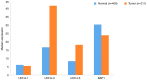
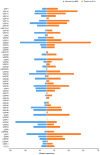
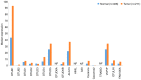
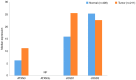
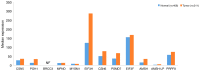
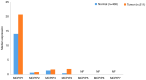
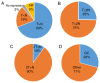
The epigenetic regulation of gene expression (via DNA methylation, histone modification and microRNA interference) contributes to a variety of diseases, particularly cancer. Protein deubiquitination serves a key role in the mechanism underlying histone modification, and consequently influences tumor development and progression. Improved characterization of the role of ubiquitinating enzymes has led to the identification of numerous deubiquitinating enzymes (DUBs) with various functions. Gastric cancer (GC) is a highly prevalent cancer type that exhibits a high mortality rate. Latest analysis about cancer patient revealed that GC is sixth deadliest cancer type, which frequently occur in male (7.2%) than female (4.1%). Complex associations between DUBs and GC progression have been revealed in multiple studies; however, the molecular mechanism underpinning the metastasis and recurrence of GC is yet to be elucidated. Generally, DUBs were upregulated in gastric cancer. The relation of DUBs and tumor size, classification and staging was observed in GC. Besides, 5-yar survival rate of patients with GC is effeccted by expression level of DUBs. Among the highly expressed DUBs, specifically six DUBs namely UCHs, USPs, OTUs, MJDs, JAMMs and MCPIPs effect on this survival rate. Consequently, the association between GC and DUBs has received increasing attention in recent years. Therefore, in the present review, literature investigating the association between DUBs and GC pathophysiology was analyzed and critically appraised.
Keywords: deubiquitinating enzymes; gastric cancer; survival.
Copyright: © Sun et al.
Figures
Similar articles
-
Targeting the signalling pathways regulated by deubiquitinases for prostate cancer therapeutics.Cell Biochem Funct. 2019 Jul;37(5):304-319. doi: 10.1002/cbf.3401. Epub 2019 May 6. Cell Biochem Funct. 2019. PMID: 31062387 Review.
-
The business of deubiquitination - location, location, location.F1000Res. 2016 Feb 11;5:F1000 Faculty Rev-163. doi: 10.12688/f1000research.7220.1. eCollection 2016. F1000Res. 2016. PMID: 26918171 Free PMC article. Review.
-
p53 stability is regulated by diverse deubiquitinating enzymes.Biochim Biophys Acta Rev Cancer. 2017 Dec;1868(2):404-411. doi: 10.1016/j.bbcan.2017.08.001. Epub 2017 Aug 8. Biochim Biophys Acta Rev Cancer. 2017. PMID: 28801249 Review.
-
Structural and Functional Basis of JAMM Deubiquitinating Enzymes in Disease.Biomolecules. 2022 Jun 29;12(7):910. doi: 10.3390/biom12070910. Biomolecules. 2022. PMID: 35883466 Free PMC article. Review.
-
Targeting deubiquitinating enzymes in cancer stem cells.Cancer Cell Int. 2017 Nov 3;17:101. doi: 10.1186/s12935-017-0472-0. eCollection 2017. Cancer Cell Int. 2017. PMID: 29142505 Free PMC article. Review.
Cited by
-
Low EBI3 Expression Promotes the Malignant Degree of Gastric Cancer.Dis Markers. 2022 Mar 17;2022:5588043. doi: 10.1155/2022/5588043. eCollection 2022. Dis Markers. 2022. PMID: 39291183 Free PMC article.
-
The Role of Ubiquitin-Proteasome System (UPS) in Asthma Pathology.J Asthma Allergy. 2025 Mar 1;18:307-330. doi: 10.2147/JAA.S490039. eCollection 2025. J Asthma Allergy. 2025. PMID: 40046173 Free PMC article. Review.
-
Physiological roles and therapeutic implications of USP6.Cell Death Discov. 2025 May 10;11(1):231. doi: 10.1038/s41420-025-02466-0. Cell Death Discov. 2025. PMID: 40348771 Free PMC article. Review.
-
Regulation of Hippo/YAP axis in colon cancer progression by the deubiquitinase JOSD1.Cell Death Discov. 2024 Aug 14;10(1):365. doi: 10.1038/s41420-024-02136-7. Cell Death Discov. 2024. PMID: 39143074 Free PMC article.
-
Activation and selectivity of OTUB-1 and OTUB-2 deubiquitinylases.J Biol Chem. 2020 May 15;295(20):6972-6982. doi: 10.1074/jbc.RA120.013073. Epub 2020 Apr 7. J Biol Chem. 2020. PMID: 32265297 Free PMC article.
References
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous
