Bioinformatics analysis to screen key genes in papillary thyroid carcinoma
- PMID: 31897130
- PMCID: PMC6924100
- DOI: 10.3892/ol.2019.11100
Bioinformatics analysis to screen key genes in papillary thyroid carcinoma
Abstract
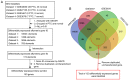
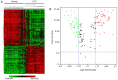
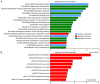
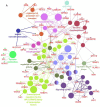
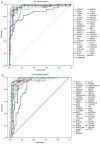
Papillary thyroid carcinoma (PTC) is the most common type of thyroid carcinoma, and its incidence has been on the increase in recent years. However, the molecular mechanism of PTC is unclear and misdiagnosis remains a major issue. Therefore, the present study aimed to investigate this mechanism, and to identify key prognostic biomarkers. Integrated analysis was used to explore differentially expressed genes (DEGs) between PTC and healthy thyroid tissue. To investigate the functions and pathways associated with DEGs, Gene Ontology, pathway and protein-protein interaction (PPI) network analyses were performed. The predictive accuracy of DEGs was evaluated using the receiver operating characteristic (ROC) curve. Based on the four microarray datasets obtained from the Gene Expression Omnibus database, namely GSE33630, GSE27155, GSE3467 and GSE3678, a total of 153 DEGs were identified, including 66 upregulated and 87 downregulated DEGs in PTC compared with controls. These DEGs were significantly enriched in cancer-related pathways and the phosphoinositide 3-kinase-AKT signaling pathway. PPI network analysis screened out key genes, including acetyl-CoA carboxylase beta, cyclin D1, BCL2, and serpin peptidase inhibitor clade A member 1, which may serve important roles in PTC pathogenesis. ROC analysis revealed that these DEGs had excellent predictive performance, thus verifying their potential for clinical diagnosis. Taken together, the findings of the present study suggest that these genes and related pathways are involved in key events of PTC progression and facilitate the identification of prognostic biomarkers.
Keywords: integrated analysis; key genes; microarray; papillary thyroid carcinoma.
Copyright: © Liu et al.
Figures


References
LinkOut - more resources
Full Text Sources
Research Materials
