Loss of Stag2 cooperates with EWS-FLI1 to transform murine Mesenchymal stem cells
- PMID: 31898537
- PMCID: PMC6941350
- DOI: 10.1186/s12885-019-6465-8
Loss of Stag2 cooperates with EWS-FLI1 to transform murine Mesenchymal stem cells
Abstract
Background: Ewing sarcoma is a malignancy of primitive cells, possibly of mesenchymal origin. It is probable that genetic perturbations other than EWS-FLI1 cooperate with it to produce the tumor. Sequencing studies identified STAG2 mutations in approximately 15% of cases in humans. In the present study, we hypothesize that loss of Stag2 cooperates with EWS-FLI1 in generating sarcomas derived from murine mesenchymal stem cells (MSCs).
Methods: Mice bearing an inducible EWS-FLI1 transgene were crossed to p53-/- mice in pure C57/Bl6 background. MSCs were derived from the bone marrow of the mice. EWS-FLI1 induction and Stag2 knockdown were achieved in vitro by adenovirus-Cre and shRNA-bearing pGIPZ lentiviral infection, respectively. The cells were then treated with ionizing radiation to 10 Gy. Anchorage independent growth in vitro was assessed by soft agar assays. Cellular migration and invasion were evaluated by transwell assays. Cells were injected with Matrigel intramuscularly into C57/Bl6 mice to test for tumor formation.
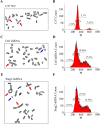
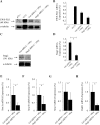
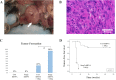
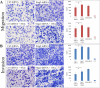
Results: Primary murine MSCs with the genotype EWS-FLI1 p53-/- were resistant to transformation and did not form tumors in syngeneic mice without irradiation. Stag2 inhibition increased the efficiency and speed of sarcoma formation significantly in irradiated EWS-FLI1 p53-/- MSCs. The efficiency of tumor formation was 91% for cells in mice injected with Stag2-repressed cells and 22% for mice receiving cells without Stag2 inhibition (p < .001). Stag2 knockdown reduced survival of mice in Kaplan-Meier analysis (p < .001). It also increased MSC migration and invasion in vitro but did not affect proliferation rate or aneuploidy.
Conclusion: Loss of Stag2 has a synergistic effect with EWS-FLI1 in the production of sarcomas from murine MSCs, but the mechanism may not relate to increased proliferation or chromosomal instability. Primary murine MSCs are resistant to transformation, and the combination of p53 null mutation, EWS-FLI1, and Stag2 inhibition does not confer immediate conversion of MSCs to sarcomas. Irradiation is necessary in this model, suggesting that perturbations of other genes beside Stag2 and p53 are likely to be essential in the development of EWS-FLI1-driven sarcomas from MSCs.
Keywords: EWS-FLI1; Ewing sarcoma; Mesenchymal stem cells; Mouse model; Stag2; p53.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Cotterill SJ, Ahrens S, Paulussen M, Jurgens HF, Voute PA, Gadner H, Craft AW. Prognostic factors in Ewing’s tumor of bone: analysis of 975 patients from the European intergroup cooperative Ewing’s sarcoma study group. J Clin Oncol. 2000;18(17):3108–3114. doi: 10.1200/JCO.2000.18.17.3108. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

