How to Illuminate the Druggable Genome Using Pharos
- PMID: 31898878
- PMCID: PMC7818358
- DOI: 10.1002/cpbi.92
How to Illuminate the Druggable Genome Using Pharos
Abstract
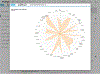
Pharos is an integrated web-based informatics platform for the analysis of data aggregated by the Illuminating the Druggable Genome (IDG) Knowledge Management Center, an NIH Common Fund initiative. The current version of Pharos (as of October 2019) spans 20,244 proteins in the human proteome, 19,880 disease and phenotype associations, and 226,829 ChEMBL compounds. This resource not only collates and analyzes data from over 60 high-quality resources to generate these types, but also uses text indexing to find less apparent connections between targets, and has recently begun to collaborate with institutions that generate data and resources. Proteins are ranked according to a knowledge-based classification system, which can help researchers to identify less studied "dark" targets that could be potentially further illuminated. This is an important process for both drug discovery and target validation, as more knowledge can accelerate target identification, and previously understudied proteins can serve as novel targets in drug discovery. Two basic protocols illustrate the levels of detail available for targets and several methods of finding targets of interest. An Alternate Protocol illustrates the difference in available knowledge between less and more studied targets. © 2020 by John Wiley & Sons, Inc. Basic Protocol 1: Search for a target and view details Alternate Protocol: Search for dark target and view details Basic Protocol 2: Filter a target list to get refined results.
Keywords: bioinformatics; dark genome; disease; drug discovery; drug targets; phenotype; proteins; target validation.
© 2020 John Wiley & Sons, Inc.
Figures







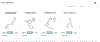






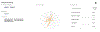







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

