A genome alignment of 120 mammals highlights ultraconserved element variability and placenta-associated enhancers
- PMID: 31899510
- PMCID: PMC6941714
- DOI: 10.1093/gigascience/giz159
A genome alignment of 120 mammals highlights ultraconserved element variability and placenta-associated enhancers
Abstract
Background: Multiple alignments of mammalian genomes have been the basis of many comparative genomic studies aiming at annotating genes, detecting regions under evolutionary constraint, and studying genome evolution. A key factor that affects the power of comparative analyses is the number of species included in a genome alignment.
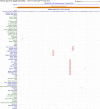
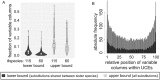
Results: To utilize the increased number of sequenced genomes and to provide an accessible resource for genomic studies, we generated a mammalian genome alignment comprising 120 species. We used this alignment and the CESAR method to provide protein-coding gene annotations for 119 non-human mammals. Furthermore, we illustrate the utility of this alignment by 2 exemplary analyses. First, we quantified how variable ultraconserved elements (UCEs) are among placental mammals. Leveraging the high taxonomic coverage in our alignment, we estimate that UCEs contain on average 4.7%-15.6% variable alignment columns. Furthermore, we show that the center regions of UCEs are generally most constrained. Second, we identified enhancer sequences that are only conserved in placental mammals. We found that these enhancers are significantly associated with placenta-related genes, suggesting that some of these enhancers may be involved in the evolution of placental mammal-specific aspects of the placenta.
Conclusion: The 120-mammal alignment and all other data are available for analysis and visualization in a genome browser at https://genome-public.pks.mpg.de/and for download at https://bds.mpi-cbg.de/hillerlab/120MammalAlignment/.
Keywords: comparative gene annotation; enhancers; genome alignment; mammals; ultraconserved elements.
© The Author(s) 2020. Published by Oxford University Press.
Figures

References
-
- Miller W, Makova KD, Nekrutenko A, et al. .. Comparative genomics. Annu Rev Genomics Hum Genet. 2004;5:15–56. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

