Mapping axon initial segment structure and function by multiplexed proximity biotinylation
- PMID: 31900387
- PMCID: PMC6941957
- DOI: 10.1038/s41467-019-13658-5
Mapping axon initial segment structure and function by multiplexed proximity biotinylation
Abstract
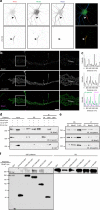
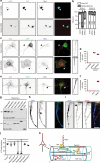
Axon initial segments (AISs) generate action potentials and regulate the polarized distribution of proteins, lipids, and organelles in neurons. While the mechanisms of AIS Na+ and K+ channel clustering are understood, the molecular mechanisms that stabilize the AIS and control neuronal polarity remain obscure. Here, we use proximity biotinylation and mass spectrometry to identify the AIS proteome. We target the biotin-ligase BirA* to the AIS by generating fusion proteins of BirA* with NF186, Ndel1, and Trim46; these chimeras map the molecular organization of AIS intracellular membrane, cytosolic, and microtubule compartments. Our experiments reveal a diverse set of biotinylated proteins not previously reported at the AIS. We show many are located at the AIS, interact with known AIS proteins, and their loss disrupts AIS structure and function. Our results provide conceptual insights and a resource for AIS molecular organization, the mechanisms of AIS stability, and polarized trafficking in neurons.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

