De novo generation of hit-like molecules from gene expression signatures using artificial intelligence
- PMID: 31900408
- PMCID: PMC6941972
- DOI: 10.1038/s41467-019-13807-w
De novo generation of hit-like molecules from gene expression signatures using artificial intelligence
Abstract
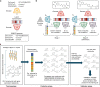
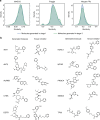
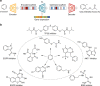
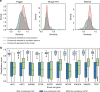
Finding new molecules with a desired biological activity is an extremely difficult task. In this context, artificial intelligence and generative models have been used for molecular de novo design and compound optimization. Herein, we report a generative model that bridges systems biology and molecular design, conditioning a generative adversarial network with transcriptomic data. By doing so, we can automatically design molecules that have a high probability to induce a desired transcriptomic profile. As long as the gene expression signature of the desired state is provided, this model is able to design active-like molecules for desired targets without any previous target annotation of the training compounds. Molecules designed by this model are more similar to active compounds than the ones identified by similarity of gene expression signatures. Overall, this method represents an alternative approach to bridge chemistry and biology in the long and difficult road of drug discovery.
Conflict of interest statement
D.A.C. and J.W. are employees of Bayer AG. O.M.L., B.B., and D.R. work directly or indirectly for Bayer SAS.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

