Sharpening the Molecular Scissors: Advances in Gene-Editing Technology
- PMID: 31901636
- PMCID: PMC6941877
- DOI: 10.1016/j.isci.2019.100789
Sharpening the Molecular Scissors: Advances in Gene-Editing Technology
Abstract
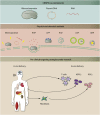
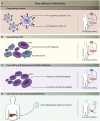
The ability to precisely modify human genes has been made possible by the development of tools such as meganucleases, zinc finger nucleases, TALENs, and CRISPR/Cas. These now make it possible to generate targeted deletions, insertions, gene knock outs, and point variants; to modulate gene expression by targeting transcription factors or epigenetic machineries to DNA; or to target and modify RNA. Endogenous repair mechanisms are used to make the modifications required in DNA; they include non-homologous end joining, homology-directed repair, homology-independent targeted integration, microhomology-mediated end joining, base-excision repair, and mismatch repair. Off-target effects can be monitored using in silico prediction and sequencing and minimized using Cas proteins with higher accuracy, such as high-fidelity Cas9, enhanced-specificity Cas9, and hyperaccurate Cas9. Alternatives to Cas9 have been identified, including Cpf1, Cas12a, Cas12b, and smaller Cas9 orthologs such as CjCas9. Delivery of gene-editing components is performed ex vivo using standard techniques or in vivo using AAV, lipid nanoparticles, or cell-penetrating peptides. Clinical development of gene-editing technology is progressing in several fields, including immunotherapy in cancer treatment, antiviral therapy for HIV infection, and treatment of genetic disorders such as β-thalassemia, sickle cell disease, lysosomal storage disorders, and retinal dystrophy. Here we review these technological advances and the challenges to their clinical implementation.
Keywords: Genetics; Molecular Biology; Techniques in Genetics.
Copyright © 2019 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests A.T.v.d.P. has provided consulting services for various industries in the field of Pompe disease under an agreement between these industries and Erasmus MC, Rotterdam, The Netherlands.
Figures


References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

