The Dynamics of Cytoplasmic mRNA Metabolism
- PMID: 31902669
- PMCID: PMC7265681
- DOI: 10.1016/j.molcel.2019.12.005
The Dynamics of Cytoplasmic mRNA Metabolism
Abstract
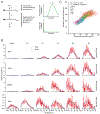
For all but a few mRNAs, the dynamics of metabolism are unknown. Here, we developed an experimental and analytical framework for examining these dynamics for mRNAs from thousands of genes. mRNAs of mouse fibroblasts exit the nucleus with diverse intragenic and intergenic poly(A)-tail lengths. Once in the cytoplasm, they have a broad (1000-fold) range of deadenylation rate constants, which correspond to cytoplasmic lifetimes. Indeed, with few exceptions, degradation appears to occur primarily through deadenylation-linked mechanisms, with little contribution from either endonucleolytic cleavage or deadenylation-independent decapping. Most mRNA molecules degrade only after their tail lengths fall below 25 nt. Decay rate constants of short-tailed mRNAs vary broadly (1000-fold) and are larger for short-tailed mRNAs that have previously undergone more rapid deadenylation. This coupling helps clear rapidly deadenylated mRNAs, enabling the large range in deadenylation rate constants to impart a similarly large range in stabilities.
Keywords: 5-ethynyl uridine; PAL- seq; TAIL-seq; deadenylation rates; decapping rates; mRNA decay rates; mRNA uridylation; metabolic labeling; poly(A)-tail lengths.
Copyright © 2019 Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of Interests The authors declare no competing interests.
Figures

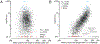
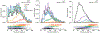
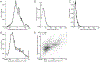


References
-
- Bönisch C, Temme C, Moritz B, and Wahle E (2007). Degradation of hsp70 and other mRNAs in Drosophila via the 5′ 3′ pathway and its regulation by heat shock. J Biol Chem 282, 21818–21828. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

