Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer
- PMID: 31902674
- PMCID: PMC7000340
- DOI: 10.1016/j.ebiom.2019.11.008
Dysregulation of the splicing machinery is directly associated to aggressiveness of prostate cancer
Abstract
Background: Dysregulation of splicing variants (SVs) expression has recently emerged as a novel cancer hallmark. Although the generation of aberrant SVs (e.g. AR-v7/sst5TMD4/etc.) is associated to prostate-cancer (PCa) aggressiveness and/or castration-resistant PCa (CRPC) development, whether the molecular reason behind such phenomena might be linked to a dysregulation of the cellular machinery responsible for the splicing process [spliceosome-components (SCs) and splicing-factors (SFs)] has not been yet explored.
Methods: Expression levels of 43 key SCs and SFs were measured in two cohorts of PCa-samples: 1) Clinically-localized formalin-fixed paraffin-embedded PCa-samples (n = 84), and 2) highly-aggressive freshly-obtained PCa-samples (n = 42).
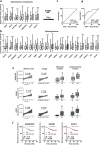
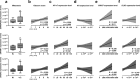
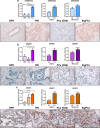
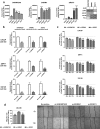
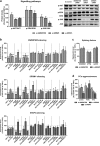
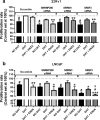
Findings: A profound dysregulation in the expression of multiple components of the splicing machinery (i.e. 7 SCs/19 SFs) were found in PCa compared to their non-tumor adjacent-regions. Notably, overexpression of SNRNP200, SRSF3 and SRRM1 (mRNA and/or protein) were associated with relevant clinical (e.g. Gleason score, T-Stage, metastasis, biochemical recurrence, etc.) and molecular (e.g. AR-v7 expression) parameters of aggressiveness in PCa-samples. Functional (cell-proliferation/migration) and mechanistic [gene-expression (qPCR) and protein-levels (western-blot)] assays were performed in normal prostate cells (PNT2) and PCa-cells (LNCaP/22Rv1/PC-3/DU145 cell-lines) in response to SNRNP200, SRSF3 and/or SRRM1 silencing (using specific siRNAs) revealed an overall decrease in proliferation/migration-rate in PCa-cells through the modulation of key oncogenic SVs expression levels (e.g. AR-v7/PKM2/XBP1s) and alteration of oncogenic signaling pathways (e.g. p-AKT/p-JNK).
Interpretation: These results demonstrate that the spliceosome is drastically altered in PCa wherein SNRNP200, SRSF3 and SRRM1 could represent attractive novel diagnostic/prognostic and therapeutic targets for PCa and CRPC.
Keywords: Prostate cancer; SNRNP200; SRRM1; SRSF3; Spliceosome; Splicing; Therapeutic target.
Copyright © 2019 The Author(s). Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of Competing Interest The authors declare that they have no conflict of interest.
Figures
References
-
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. - PubMed
-
- Hormaechea-Agulla D., Jimenez-Vacas J.M., Gomez-Gomez E., F L.L., Carrasco-Valiente J., Valero-Rosa J. The oncogenic role of the spliced somatostatin receptor sst5TMD4 variant in prostate cancer. Faseb J. 2017;31(11):4682–4696. - PubMed
-
- Wong N., Yan J., Ojo D., De Melo J., Cutz J.C., Tang D. Changes in PKM2 associate with prostate cancer progression. Cancer Invest. 2014;32(7):330–338. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

