Macrophage MSR1 promotes BMSC osteogenic differentiation and M2-like polarization by activating PI3K/AKT/GSK3β/β-catenin pathway
- PMID: 31903103
- PMCID: PMC6929615
- DOI: 10.7150/thno.36930
Macrophage MSR1 promotes BMSC osteogenic differentiation and M2-like polarization by activating PI3K/AKT/GSK3β/β-catenin pathway
Abstract
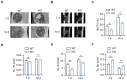
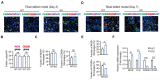
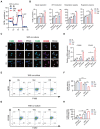
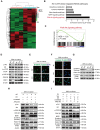
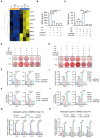
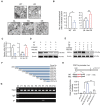
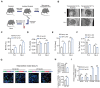
Approximately 10% of bone fractures do not heal satisfactorily, leading to significant clinical and socioeconomic implications. Recently, the role of macrophages in regulating bone marrow stem cell (BMSC) differentiation through the osteogenic pathway during fracture healing has attracted much attention. Methods: The tibial monocortical defect model was employed to determine the critical role of macrophage scavenger receptor 1 (MSR1) during intramembranous ossification (IO) in vivo. The potential functions and mechanisms of MSR1 were explored in a co-culture system of bone marrow-derived macrophages (BMDMs), RAW264.7 cells, and BMSCs using qPCR, Western blotting, immunofluorescence, and RNA sequencing. Results: In this study, using the tibial monocortical defect model, we observed delayed IO in MSR1 knockout (KO) mice compared to MSR1 wild-type (WT) mice. Furthermore, macrophage MSR1 mediated PI3K/AKT/GSK3β/β-catenin signaling increased ability to promote osteogenic differentiation of BMSCs in the co-culture system. We also identified proliferator-activated receptor gamma coactivator 1-alpha (PGC1α) as the target gene for macrophage MSR1-activated PI3K/AKT/GSK3β/β-catenin pathway in the co-culture system that facilitated M2-like polarization by enhancing mitochondrial oxidative phosphorylation. Conclusion: Our findings revealed a previously unrecognized function of MSR1 in macrophages during fracture repair. Targeting MSR1 might, therefore, be a new therapeutic strategy for fracture repair.
Keywords: Bone marrow stem cells; Macrophage scavenger receptor 1; Osteogenic differentiation; Oxidative phosphorylation; PI3K/AKT/GSK3β/β-catenin pathway.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures



References
-
- Hak DJ, Fitzpatrick D, Bishop JA, Marsh JL, Tilp S, Schnettler R. et al. Delayed union and nonunions: epidemiology, clinical issues, and financial aspects. Injury. 2014;45(Suppl 2):S3–7. - PubMed
-
- Gerstenfeld LC, Cullinane DM, Barnes GL, Graves DT, Einhorn TA. Fracture healing as a post-natal developmental process: molecular, spatial, and temporal aspects of its regulation. J Cell Biochem. 2003;88:873–84. - PubMed
-
- Schindeler A, McDonald MM, Bokko P, Little DG. Bone remodeling during fracture repair: The cellular picture. Semin Cell Dev Biol. 2008;19:459–66. - PubMed
-
- Thompson Z, Miclau T, Hu D, Helms JA. A model for intramembranous ossification during fracture healing. J Orthop Res. 2002;20:1091–8. - PubMed
-
- Phillips AM. Overview of the fracture healing cascade. Injury. 2005;36(Suppl 3):S5–7. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

